Abstract
Genetic control of the timing of flowering in woody plants is complex and has yet to be adequately investigated due to their long life-cycle and difficulties in genetic modification. Studies in Populus, one of the best woody plant models, have revealed a highly conserved genetic network for flowering timing in annuals. However, traits like continuous flowering cannot be addressed with Populus. Roses and strawberries have relatively small, diploid genomes and feature enormous natural variation. With the development of new genetic populations and genomic tools, roses and strawberries have become good models for studying the molecular mechanisms underpinning the regulation of flowering in woody plants. Here, we review findings on the molecular and genetic factors controlling continuous flowering in roses and woodland strawberries. Natural variation at TFL1 orthologous genes in both roses and strawberries seems be the key plausible factor that regulates continuous flowering. However, recent efforts suggest that a two-recessive-loci model may explain the controlling of continuous flowering in roses. We propose that epigenetic factors, including non-coding RNAs or chromatin-related factors, might also play a role. Insights into the genetic control of flowering time variation in roses should benefit the development of new germplasm for woody crops and shed light on the molecular genetic bases for the production and maintenance of plant biodiversity.
Keywords: Rose, Continuous flowering, Model woody plant, Genetics, Bulk-segregation analysis, Genome-wide prediction
Understanding the molecular mechanisms underlying plant adaptation to local environmental conditions has been a long-standing goal in biology. Timing of floral transition, i.e., the switch from vegetative to reproductive growth, determines the reproductive success and ultimate fitness for plant adaptation. In annual and herbaceous model plants like Arabidopsis and rice, the molecular genetic networks controlling floral transition have been extensively investigated. However, the regulation of flowering time in perennial woody plants has just begun to be elucidated, especially in Populus (Brunner et al., 2014, Ding and Nilsson, 2016, Jansson and Douglas, 2007, Wang et al., 2011). Due to their perennial and long life-history, woody plants feature specific strategies, such as bud dormancy, to adapt to the ever-changing environmental conditions in which they have to grow for many years. Populus has been used as a model species in woody plant biology (Ellis et al., 2010). It was the first tree species to have its genome completely sequenced (Tuskan et al., 2006). In addition, high transformation rates in Populus allow for functional analysis of genes (Fillatti et al., 1987). Using genetic and molecular approaches, researchers have found that the genetic basis controlling floral transition is highly conserved between Populus and Arabidopsis. Several reviews have summarized recent progress in Populus (Brunner et al., 2014, Ding and Nilsson, 2016, Petterle et al., 2013). Here we will mainly focus on genetic studies of the control of flowering time in roses and related species.
1. Roses as a model woody plant
1.1. Roses are important plants for human beings
Rosoideae plants (including roses, strawberries, red and black raspberries, blackberries, etc.) are of great economic importance, and thus receive much attention not only from scientists but also from breeders. As one of the most popular ornamental plants, roses have great cultural value related to continuous flowering, scent and color. Besides the unprecedented importance in horticulture, roses are important materials for cut flowers, production of essential oils in perfume and cosmetic products, and even for food production. For example, roses are key ingredients in flower cake, a highly popular dessert, which was first recorded in the Qing Dynasty in Yunnan, China. In addition, roses have been widely used for medicinal purposes for many years. Taken together, roses represent a very important crop for human beings.
1.2. Roses feature many important traits, including continuous flowering
Roses have important biological traits like flowering, flower architecture, flower color and scent, easy vegetative propagation and more. Some of these biological traits are also key economic characteristics of roses. Continuous flowering is such a trait. The complex but well-documented history of rose domestication mainly involves hybridization among different species of the Rosa plants (see review by Bendahmane et al. (2013)). The Chinese rose, Rosa chinensis ‘Old Blush’, contributes many key traits to current cultivated roses (Rosa x hybrida). One of these important traits is recurrent blooming (RB), also known as continuous flowering phenotype (CF; flowers continuously during the year), in contrast to the once-flowering phenotype (OF; flowering only a single period of a year). Guaranteeing a constant supply of raw materials for cut flowers, fruits and other related products, continuous flowering is a key biological trait of both roses and strawberries (Fragaria) that is economically important. Therefore, many scientists have endeavored to understand the molecular genetic control of this trait.
1.3. Some roses have a short life cycle
Like many shrub plants, many wild species of perennial roses start floral transition either in late autumn or early spring but flower in spring when day length and temperature are acceptable. These species normally only flower once a year, while prior to flowering they normally need several months/years of vegetative growth. However, some roses have a very short juvenile phase and start reproductive growth shortly after seed germination (Duchesne, 1766, Vries, 1976). R. chinensis ‘Old Blush’ (Hurst, 1941) represents such a case: it starts to flower with only 6–8 weeks of juvenile phase (post seed germination; Fig. 1) (Li et al., 2015b), similar to the flowering time of Arabidopsis thaliana Col-0 under long-day conditions (Andres and Coupland, 2012, Fornara et al., 2010, Hu et al., 2014). This short juvenile-phase makes rose species a suitable model species of woody plants.
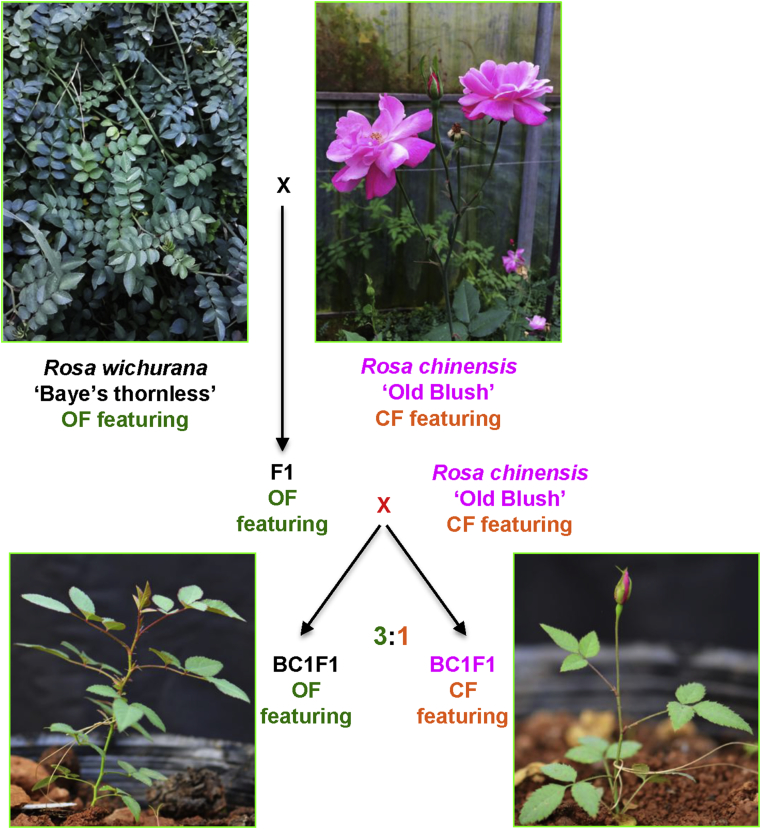
Fig. 1.
The construction of a new mapping population for continuous flowering (CF) with R. wichurana ‘Baye's thornless’ and R. chinensis ‘Old Blush’ as parents. R. wichurana and R. chinensis feature OF (flowering once a year) and CF behaviors, respectively. F1s between these two parents show the OF phenotype indicating the recessive nature of CF-controlling gene(s). The backcross between F1s and CF-featuring R. chinensis generates 3:1 segregating (OF to CF featuring) BC1F1s. CF-featuring BC1F1 can flower about six weeks after germination, while the OF-featuring plants need at least one year of vegetative growth prior to blooming (modified according to Li et al., 2015b).
1.4. Diploid nature of some roses
Rosa species feature highly variable ploidy levels, ranging from diploids to polyploids (Li et al., 2015b, Roberts et al., 2009, Vamosi and Dickinson, 2006, Yokoya et al., 2000). Rosa prealucens from the Sino-Himalayan region seems to even be a decaploid, the highest naturally occurring ploidy level in Rosa (Jian et al., 2010). Interestingly, the old garden rose R. chinensis ‘Old Blush’ and Rosa wichurana ‘Baye's thornless' are diploid species, with genome sizes about 600–800 Mb, five to six times that of A. thaliana, or three times the size of Fragaria vesca (Longhi et al., 2014, Yokoya et al., 2000). A comparatively small, diploid genome makes whole-genome sequencing relatively feasible. The F. vesca genome (diploid; about 200 Mb) is the first genome that has been sequenced with relatively high quality in the Rosoideae family Rosaceae (Shulaev et al., 2011). Currently the ‘Rose Genome Sequence Initiative’ is sequencing the R. chinensis genome with NGS technologies combined with traditional molecular marker data (Foucher et al., 2015). However, genome heterozygosity is always a problem in Rosa species due to self-incompatibility and frequent inter-species hybridization (Foucher et al., 2015).
1.5. Available genomic and genetic tools/resources
Many molecular genetic and genomic resources and tools have been developed over the past decades (please see review by Longhi et al. (2014)). For roses, many mapping populations have been created via crossing different species/cultivars to reveal the genetic mechanisms underlying various traits like flower color, petal number, continuous flowering and scent (Crespel et al., 2002, Debener and Mattiesch, 1999, Debener et al., 2001, Dugo et al., 2005, Li et al., 2015b, Linde et al., 2006, Moghaddam et al., 2012, Spiller et al., 2011, Yan et al., 2007). One genetic linkage-map for tetraploid roses was even successfully constructed (Yu et al., 2015a). However, these populations are normally relatively small (about 100 individuals), and not large enough to map ideal candidate genes to a small and precise region. Interestingly, with a bigger F1 population (created by crossing diploid R. wichurana and dihaploid Rosa hybrida) containing about 670 individuals, Iwata et al. (2012) found RoKSN, one of the key regulators of continuous flowering in both roses and strawberries. Recently, a new rose mapping population, containing about 800 lines (including parents, F1s, and BC1F1s), was created by crossing the diploids R. chinensis ‘Old Blush’ and R. wichurana ‘Basye's thornless'. This population features at least six pairs of contrasting traits including continuous flowering (Li et al., 2015b). With this population, the loci controlling continuous flowering in R. chinensis could be identified.
1.6. Genetic transformation of roses
A good model plant species requires a stable and efficient transformation system. In Rosoideae, such transformation systems have been developed for some species, for example F. vesca. With specifically designed protocols 15–100% transformation efficiency has been achieved with certain F. vesca accessions (Alsheikh et al., 2002, Oosumi et al., 2005). It is very easy to propagate roses with apical buds or nodal segments in vitro, and plant regeneration through tissue culture methods is relatively simple (see review by Pati et al., 2006). Mainly based on tissue culture techniques, transformation experiments have been successfully carried out in both tetraploid and diploid roses. Beta-glucuronidase (GUS) (Li et al., 2002) and Ace-AMP1, an antimicrobial protein gene (Li et al., 2003), have been transformed into the tetraploid R. hybrida cv. Carefree Beauty. Other examples of successful transformation into tetraploid roses include an flavonoid 3′,5′-hydoxylase (Katsumoto et al., 2007), an ethylene-responding gene (Zakizadeh et al., 2012), and the RoKSN gene (Randoux et al., 2014), which were introduced into different cultivars of R. hybrida. Successful transformation of diploid roses has been reported relatively recently in R. chinensis (Vergne et al., 2009) and Rosa rugosa (Xing et al., 2014a, Xing et al., 2014b). It is noteworthy that the transformation efficiency was 2–15%. In general, the success of rose transformation in both diploid and tetraploid species enables the functional evaluation of important genes in combination with analogous systems like Arabidopsis (Otagaki et al., 2015) or Nicotiana benthamiana (Magnard et al., 2015). Importantly, recently developed virus-induced gene silencing (VIGS) in roses is likely to be another important tool to rapidly study gene function (Ito et al., 2012, Magnard et al., 2015).
Thus, roses represent a perennial woody model featuring highly diverse morphological traits with a widespread growth range. Robust in vitro propagation, the feasibility of transformation or genetic modifications, and a genome in the process of being sequenced currently make roses an emerging model for woody perennial plants. Furthermore, a short juvenile phase accompanied by continuous flowering has made roses a special model for studying those traits that cannot be addressed with annual species, such as A. thaliana, or with other perennial species, e.g. Populus.
2. Current status of floral transition study in roses
As woody perennial plants, the regulatory mechanisms of rose flowering receive much less attention than model annual herbaceous species like Arabidopsis or rice. Although many genes from Arabidopsis or rice can be identified or predicted (Remay et al., 2009), the functional significance of these genes awaits critical experimental evaluation to build up the gene-regulatory-networks for rose. Currently, the Arabidopsis TFL1 gene in rose (RoKSN) is the only gene related to flowering time regulation which has been functionally characterized.
2.1. TFL1 homologs are important regulators of rose flowering
The study of flowering time in roses has a long history. Semeniuk (1971) first proposed that one homozygous recessive locus, RB (for recurrent flowering), controlled the CF behavior in roses about 35 years after Cajlacjan proposed the hypothesis of florigen (Cajlacjan, 1937). However, only until 2002 Crespel et al. (2002) mapped the RB gene and found it to co-localize with RoSPINDLY, an Arabidopsis gene involved in gibberellin signaling, although these findings have not been replicated with larger populations (Spiller et al., 2011). In 2012 and 2014, the Foucher lab used QTL mapping combined with candidate gene approaches, transformation, as well as association genetic methods to show that rose RoKSN or strawberry FvKSN, putative orthologs of Arabidopsis TERMINAL FLOWER LIKE 1 (TFL1), were very likely underlying the CF phenotype (Iwata et al., 2012, Randoux et al., 2014). Similar to Arabidopsis TFL1 (Andres and Coupland, 2012, Fornara et al., 2010), through competing for RoFD with RoFT, RoKSN regulates flowering via modulating accumulation of floral integrators like RoFT, RoAP1, RoLFY, and RoSOC1 (Randoux et al., 2014). Interestingly, the TFL1 ortholog (PtTFL1) is also very important for the onset of flowering in Poplus tremuloides (Mohamed et al., 2010). Therefore, the TFL1-mediated regulation of flowering seems to be conserved in woody plants.
2.2. Natural variation in TFL1 orthologs associates with continuous flowering
In A. thaliana, natural variation at key flowering genes like FRIGIDA and FLC mediates the prominent differences of flowering time among accessions (Johanson et al., 2000, Michaels and Amasino, 1999, Michaels and Amasino, 2001, Swarup et al., 1999). Similarly, this phenomenon also occurs in woody plants. In roses featuring CF behavior, a retrotransposon is present in the second intron of RoKSN, while OF roses do not have such an insertion (Iwata et al., 2012). Interestingly, this insertion in CF roses is correlated with the repression of full length RoKSN mRNA accumulation, which is normally expressed in OF roses. Coincidently, woodland strawberries, which have the CF flowering phenotype, also have a mutation in the FvKSN gene, although the type of mutation is completely different from RoKSN; FvKSN has a two-nucleotide deletion in the first exon, which causes a premature stop codon (Iwata et al., 2012). This deletion is correlated with the CF phenotype in many varieties of F. vesca. Furthermore, over-expressing the RoKSN allele from the OF R. wichurana in CF R. hybrida causes a loss-of-CF phenotype, thus indicating that mutation in the RoKSN gene very likely controls the variation of flowering behavior in roses (Randoux et al., 2014). However, one should be cautious with this conclusion because overexpressing a transcription factor might cause artifacts (Li and Millar, 2013). Another point should be addressed is that the expression of RoKSN responds to fluctuations in GA concentrations (Randoux et al., 2012). This agrees well with results from Arabidopsis and rice that different pathways are inter-connected in the genetic control of flowering time (Andres and Coupland, 2012, Fornara et al., 2010).
Currently, rose and strawberry TFL1 orthologs remain the only candidate genes responsible for continuous flowering in both roses and strawberry. However, whether RoKSN is the only QTL underlining the CF trait in Rosa still needs further investigation.
2.3. Two loci act in regulating CF phenotype in roses
It is worth noting that the timing of flowering in roses could also be controlled by two QTL (Dugo et al., 2005). With a population containing 96 F1 individuals created by an inter-specific cross between diploid CF R. chinensis ‘Blush Noisette’ (D10) and OF R. wichurana (E15), two QTL explaining more than 70% of flowering variation (from the onset date of D10), were detected (Dugo et al., 2005). These findings indicate that the CF trait may be controlled by more than one regulator (Bendahmane et al., 2013). Interestingly, some very recent efforts provide evidence supporting this hypothesis.
In 2015, Li et al. (2015b) reported a mapping population constructed by crossing R. chinensis ‘Old Blush’ (OB) with R. wichurana ‘Basye's thornless' (W), the same species from Dugo et al. (2005) but with different accessions. This population contains more than 800 individual plants including six sub-populations consisting of parents (OB and W), F1s, F2s and two backcrosses (BC1W and BC1OB, meaning backcrossed to W and OB, respectively). All the plants in F1 (296 plants) and BC1W (150 individuals) sub-populations only flowered once a year, i.e., these plants were OF, indicating that the CF trait is controlled by recessive alleles while the two parental lines harbor homozygous states at the CF genes. Interestingly, the small F2 sub-population (with only 41 individuals) showed segregation in flowering behavior with five flowering continuously (segregating at about 1:15). The authors further explored the flowering behavior in the BC1OB sub-population. Both batches of BC1OB (141 and 159 individuals for year 2013 and 2014, respectively) showed a 1:3 segregation ratio for the CF phenotype, suggesting that the CF trait in R. chinensis is very likely controlled by two recessive homozygous loci (Fig. 1).
In summary, although TFL1 orthologs in rose and woodland strawberry seem to be responsible for the CF trait, new efforts with bigger and more complex mapping populations suggest that the biologically and economically important CF phenotype may be controlled by two or possibly more genetic loci.
3. Future research directions
3.1. Possibility of epigenetic control in rose flowering time
The past decades have witnessed enormous progress and tool development for studying epigenetic control of many biological processes. Epigenetic signals modify chromatin accessibility for transcription machinery and hence are essential for phenotypic variation (Feng and Jacobsen, 2011). Epigenetic modifications normally reflect the environmental and developmental plasticity of gene expression, hence the epigenetic influences on plant adaptive traits have attracted extensive attention (see reviews Bossdorf et al., 2008, Richards, 2011). Interestingly, epigenetic studies at FLOWERING LOCUS C (FLC), one key player in vernalization response of A. thaliana, have become model cases in plant epigenetics research (Angel et al., 2015, Coustham et al., 2012, Heo and Sung, 2011, Ietswaart et al., 2012, Li et al., 2015a, Liu et al., 2010, Marquardt et al., 2014, Song et al., 2012, Sun et al., 2013, Swiezewski et al., 2009). However, orthologous sequences of FLC have not been found in rose ESTs or RNA-seq datasets (Dubois et al., 2012, Foucher et al., 2008, Remay et al., 2009). Since temperature is a key factor regulating the flowering of OF roses such as R. wichurana (Hess et al., 2007, Kurokura et al., 2013), the molecular mechanisms underlying the transduction of environmental triggers, such as temperature, in flowering time control of roses merit further investigation. It will also be interesting to test the response of RoKSN expression to low or ambient temperature fluctuations since FvTFL1 repression of flowering in strawberries clearly depends on temperature (Nakano et al., 2015).
The insertion of a transposon in the RoKSN gene of CF roses deserves special attention (Iwata et al., 2012). Transposon insertion might result in the spread of epigenetic modifications, causing neighboring genes to be silenced (Dong et al., 2012, Verhoeven and Van Gurp, 2012). A detailed comparison of the epigenetic changes on RoKSN (like histone methylation, small-interfering RNAs or DNA methylation) in both CF and OF roses should shed light on whether epigenetic silencing is a major factor in RoKSN-mediated CF control.
It is well established that miRNAs play essential roles in regulating the timing of floral transition in both monocot and dicot plants. The miR156-SPLs module participates in the aging pathway, hence changing the period of juvenile growth to control the timing of flowering in both annual and perennial plants (see reviews Andres and Coupland, 2012, Golembeski and Imaizumi, 2015, Yu et al., 2015b, Zhou and Wang, 2013). As CF in both roses and strawberries is always accompanied by a short period of juvenile growth, the accumulation of miR156 in CF and OF roses should be determined. MiR156 belongs to the conserved miRNAs regulating key life-history traits (Cho et al., 2012, Chuck et al., 2007, Lauter et al., 2005, Nair et al., 2010, Zhu et al., 2009), while non-conserved and family/species-specific miRNAs, for example the Brasicaceae-specific miR824 (Hu et al., 2014), may also participate in regulating floral transition. Currently, no miRNA profiling data during floral transition is available for roses, although studies have evidenced the important contribution of miR164 in regulating petal growth (Pei et al., 2013a, Pei et al., 2013b). To reveal miRNA-mediated control of floral transition in roses, profiling small RNAs across developmental stages should be helpful.
4. Genomics and genetics for dissecting the molecular genetic bases of rose flowering traits
4.1. High quality reference genome of roses
A high quality reference genome is necessary for deciphering the key biological traits in roses. Currently, the ‘Rose Genome Sequence Initiative’ is working to provide a high quality genome sequence for R. chinensis ‘Old Blush’ (Foucher et al., 2015). Due to the high heterozygosity of roses, it will not be easy for researchers to assemble and construct such a map. Although the third-generation high-throughput sequencing platforms, like single-molecular real-time sequencing from Pacific Biosciences, can generate much longer reads than normal Illumina short reads, high error rate per reads remains a big challenge. However, the short-reads platforms also have their inherent, biased and non-random sequencing errors. Therefore, a good sequencing strategy, for example combining the second- and third-generation sequencing platforms together with the already-established genetic maps and markers, followed by strong bioinformatics analysis should be established. We expect that a high quality reference genome will benefit the efforts in finding out the casual gene(s) regulating key traits of roses via either mapping-by-sequencing or genome-wide prediction methods (see below).
4.2. The bulk-segregating analysis (BSA) strategy
For many decades, the powerful and successful forward-genetics approach to find/map causal gene(s) in various species has depended heavily on developing a complex and large mapping population. Today, next-generation sequencing-based mapping-by-sequencing technology allows rapid identification of gene(s) underlying specific traits at a single-nucleotide resolution even in complex genetic backgrounds (Schneeberger, 2014). One such technology is called bulk-segregating association analysis (BSA), which works via sequencing pooled individuals through whole genome sequencing. The central idea of this method relies on counting the allele frequency associated with the targeted phenotype in segregating F2 or BC1F1 populations, while high throughput sequencing is used for producing a sufficient number of markers (Schneeberger, 2014). This method has been successfully introduced in Arabidopsis (Lister et al., 2009, Schneeberger et al., 2009) and barley (Pankin et al., 2014). As the rose BC1OB sub-population shows a clear three-to-one segregation for flowering behavior (Li et al., 2015b), it should be plausible to compare the homozygous SNPs between pools (OF- and CF-featuring) of the BC1OB population to their respective parents and find the polymorphisms correlated with the CF trait in roses.
It should be pointed out that the BSA approach is suitable for finding causal mutations for not only the continuous flowering trait but also for any trait with a good mapping population and genetic investigation (Schneeberger, 2014). Besides the flowering behavior difference, plants in the newly developed population derived from a cross between R. chinensis and R. wichurana also differed in five additional traits including the presence of prickles on the stem, sensitivity to black spot, erect or prostrate growth, petal number and color (Li et al., 2015b). Since a reference genome is not a prerequisite for the BSA approach (Schneeberger, 2014), this method will be very helpful for dissecting economically important traits in many Rosoideae species including roses.
4.3. Genome-wide prediction (GWP) method
Another way to reveal the molecular genetic bases for important phenotypes of Rosoideae plants is genome-wide prediction (GWP), which predicts the expected trait value of a genotype based on dense molecular markers covering the whole genome (Meuwissen et al., 2001; see also reviews Desta and Ortiz, 2014, Falke et al., 2013). GWP estimates the effect of all markers but not the size of effect; hence this concept has been adopted widely in many animals and crops (Desta and Ortiz, 2014, Falke et al., 2013). As the genome sequences (Foucher et al., 2015, Longhi et al., 2014, Shulaev et al., 2011) and integrated genetic maps (Spiller et al., 2011) for roses and strawberry become available, many important but complex traits of these plants are expected to be investigated systematically soon.
In summary, featuring phenotypes like continuous flowering, scent production and emission, high morphological diversity, etc., roses represent an economically and biologically important model for woody plants. With the development of new mapping populations and new genomic tools, it is now feasible to unravel the molecular genetic mechanisms underlying these important traits, which cannot be addressed by using model species like Arabidopsis, rice, or even Populus. Revealing these mechanisms in roses and related species certainly will not only help breeders to develop new germplasm for woody plants, but also benefit biologists in disentangling the molecular genetics and evolution of biodiversity.
Acknowledgements
We apologize to colleagues whose work or original publications could not be included owing to space constraints. Work in the Hu lab is supported by grants from the Chinese Academy of Sciences under the “Hundreds of Talents” plan and a grant from the “Yunnan Recruitment Program of Experts in Sciences”. No conflict of interest declared.
(Editor: Xuewen Wang)
Footnotes
Peer review under responsibility of Editorial Office of Plant Diversity.
References
- Alsheikh M., Suso H.-P., Robson M., Battey N., Wetten A. Appropriate choice of antibiotic and Agrobacterium strain improves transformation of antibiotic-sensitive Fragaria vesca and F. v. semperflorens. Plant Cell Rep. 2002;20:1173–1180. [Google Scholar]
- Andres F., Coupland G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- Angel A., Song J., Yang H., Questa J.I., Dean C., Howard M. Vernalizing cold is registered digitally at FLC. Proc. Natl. Acad. Sci. U. S. A. 2015;112:4146–4151. doi: 10.1073/pnas.1503100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane M., Dubois A., Raymond O., Bris M.L. Genetics and genomics of flower initiation and development in roses. J. Exp. Bot. 2013;64:847–857. doi: 10.1093/jxb/ers387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossdorf O., Richards C.L., Pigliucci M. Epigenetics for ecologists. Ecol. Lett. 2008;11:106–115. doi: 10.1111/j.1461-0248.2007.01130.x. [DOI] [PubMed] [Google Scholar]
- Brunner A.M., Evans L.M., Hsu C.-Y., Sheng X. Vernalization and the chilling requirement to exit bud dormancy: shared or separate regulation? Front. Plant Sci. 2014;5 doi: 10.3389/fpls.2014.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajlacjan M.C. Concerning the hormonal nature of plant development processes. Comptes Rendus De. L Acad. Des. Sci. De. L Urss. 1937;16:227–230. [Google Scholar]
- Cho S.H., Coruh C., Axtell M.J. miR156 and miR390 regulate tasiRNA accumulation and developmental timing in physcomitrella patens. Plant Cell. 2012;24:4837–4849. doi: 10.1105/tpc.112.103176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Meeley R., Irish E., Sakai H., Hake S. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat. Genet. 2007;39:1517–1521. doi: 10.1038/ng.2007.20. [DOI] [PubMed] [Google Scholar]
- Coustham V., Li P., Strange A., Lister C., Song J., Dean C. Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science. 2012;337:584–587. doi: 10.1126/science.1221881. [DOI] [PubMed] [Google Scholar]
- Crespel L., Chirollet M., Durel C., Zhang D., Meynet J., Gudin S. Mapping of qualitative and quantitative phenotypic traits in Rosa using AFLP markers. Theor. Appl. Genet. 2002;105:1207–1214. doi: 10.1007/s00122-002-1102-2. [DOI] [PubMed] [Google Scholar]
- Debener T., Mattiesch L. Construction of a genetic linkage map for roses using RAPD and AFLP markers. Theor. Appl. Genet. 1999;99:891–899. [Google Scholar]
- Debener T., Mattiesch L., Vosman B. A molecular marker map for roses. In: Zieslin N., Agbaria H., editors. Proceedings of the Third International Symposium on Rose Research and Cultivation. 2001. pp. 283–287. (Acta Horticulturae; vol.) [Google Scholar]
- Desta Z.A., Ortiz R. Genomic selection: genome-wide prediction in plant improvement. Trends Plant Sci. 2014;19:592–601. doi: 10.1016/j.tplants.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Ding J., Nilsson O. Molecular regulation of phenology in trees — because the seasons they are a-changin'. Curr. Opin. Plant Biol. 2016;29:73–79. doi: 10.1016/j.pbi.2015.11.007. [DOI] [PubMed] [Google Scholar]
- Dong X., Reimer J., Gobel U. Natural variation of H3K27me3 distribution between two Arabidopsis accessions and its association with flanking transposable elements. Genome Biol. 2012;13:R117. doi: 10.1186/gb-2012-13-12-r117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois A., Carrere S., Raymond O. Transcriptome database resource and gene expression atlas for the rose. BMC Genomics. 2012;13:638. doi: 10.1186/1471-2164-13-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchesne N. Didot Panckoucke, C. J; Paris: 1766. Histoire Naturelle Des Fraisiers. [Google Scholar]
- Dugo M.L., Satovic Z., Millan T. Genetic mapping of QTLs controlling horticultural traits in diploid roses. Theor. Appl. Genet. 2005;111:511–520. doi: 10.1007/s00122-005-2042-4. [DOI] [PubMed] [Google Scholar]
- Ellis B., Jansson S., Strauss S., Tuskan G. Why and how Populus became a “model tree”. In: Jansson S., Bhalerao R., Groover A., editors. Genetics and Genomics of Populus. vol. 8. Springer; New York: 2010. pp. 3–14. (Plant Genetics and Genomics: Crops and Models). [Google Scholar]
- Falke K.C., Glander S., He F., Hu J., De Meaux J., Schmitz G. The spectrum of mutations controlling complex traits and the genetics of fitness in plants. Curr. Opin. Genet. Dev. 2013;23:665–671. doi: 10.1016/j.gde.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Feng S., Jacobsen S.E. Epigenetic modifications in plants: an evolutionary perspective. Curr. Opin. Plant Biol. 2011;14:179–186. doi: 10.1016/j.pbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatti J.J., Sellmer J., Mccown B., Haissig B., Comai L. Agrobacterium mediated transformation and regeneration of Populus. Mol. General Genet. 1987;206:192–199. [Google Scholar]
- Fornara F., De Montaigu A., Coupland G. SnapShot: control of flowering in Arabidopsis. Cell. 2010;141(550):e1–e2. doi: 10.1016/j.cell.2010.04.024. [DOI] [PubMed] [Google Scholar]
- Foucher F., Chevalier M., Corre C., Soufflet-Freslon V., Legeai F., Oyant L.H.S. New resources for studying the rose flowering process. Genome. 2008;51:827–837. doi: 10.1139/G08-067. [DOI] [PubMed] [Google Scholar]
- Foucher F., Hibrand-Saint Oyant L., Hamama L. Towards the rose genome sequence and its use in research and breeding. In: Debener T., Linde M., editors. Vi International Symposium on Rose Research and Cultivation. vol. 1064. Leuven 1: Int Soc Horticultural Science; 2015. pp. 167–175. (Acta Horticulturae). [Google Scholar]
- Golembeski G.S., Imaizumi T. Photoperiodic regulation of florigen function in Arabidopsis thaliana. Arabidopsis Book/Am. Soc. Plant Biol. 2015;13:e0178. doi: 10.1199/tab.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J.B., Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- Hess G., Byrne D.H., Zhang H.B. Toward positional cloning of everblooming gene (evb) in plants: a BAC library of Rosa chinensis cv. Old Blush. In: Pemberton H.B., editor. Proceedings of the IVth International Symposium on Rose Research and Cultivation. Leuven 1: International Society Horticultural Science; 2007. pp. 169–174. (Acta Horticulturae; vol.) [Google Scholar]
- Hu J.Y., Zhou Y., He F. miR824-Regulated AGAMOUS-LIKE16 contributes to flowering time repression in Arabidopsis. Plant Cell. 2014;26:2024–2037. doi: 10.1105/tpc.114.124685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst C.C. Notes on the origin and evolution of our garden roses. J. R. Hortic. Soc. 1941;66:73–82. [Google Scholar]
- Ietswaart R., Wu Z., Dean C. Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 2012;28:445–453. doi: 10.1016/j.tig.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Ito H., Ochiai M., Kato H. Rose phytoene desaturase gene silencing by apple latent spherical virus vectors. HortScience. 2012;47:1278–1282. [Google Scholar]
- Iwata H., Gaston A., Remay A. The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J. 2012;69:116–125. doi: 10.1111/j.1365-313X.2011.04776.x. [DOI] [PubMed] [Google Scholar]
- Jansson S., Douglas C.J. Annual Review of Plant Biology. vol. 58. Palo Alto: Annual Reviews; 2007. Populus: a model system for plant biology; pp. 435–458. (Annual Review of Plant Biology). [DOI] [PubMed] [Google Scholar]
- Jian H.Y., Zhang H., Tang K.X. Decaploidy in Rosa praelucens byhouwer (Rosaceae) endemic to Zhongdian Plateau, Yunnan, China. Caryologia. 2010;63:162–167. [Google Scholar]
- Johanson U., West J., Lister C., Michaels S., Amasino R., Dean C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- Katsumoto Y., Fukuchi-Mizutani M., Fukui Y. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 2007;48:1589–1600. doi: 10.1093/pcp/pcm131. [DOI] [PubMed] [Google Scholar]
- Kurokura T., Mimida N., Battey N.H., Hytonen T. The regulation of seasonal flowering in the Rosaceae. J. Exp. Bot. 2013;64:4131–4141. doi: 10.1093/jxb/ert233. [DOI] [PubMed] [Google Scholar]
- Lauter N., Kampani A., Carlson S., Goebel M., Moose S.P. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9412–9417. doi: 10.1073/pnas.0503927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Millar A.A. A microRNA-resistant target transgene generates artefacts that misrepresent endogenous microRNA function in Arabidopsis. Mol. Plant. 2013;6:577–580. doi: 10.1093/mp/sss136. [DOI] [PubMed] [Google Scholar]
- Li P., Tao Z., Dean C. Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes & Dev. 2015;29:696–701. doi: 10.1101/gad.258814.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhou N., Zhou Q. Inheritance of perpetual blooming in Rosa chinensis ‘Old Blush’. Hortic. Plant J. 2015;1:108–112. [Google Scholar]
- Li X., Gasic K., Cammue B., Broekaert W., Korban S.S. Transgenic rose lines harboring an antimicrobial protein gene, Ace-AMP1, demonstrate enhanced resistance to powdery mildew (Sphaerotheca pannosa) Planta. 2003;218:226–232. doi: 10.1007/s00425-003-1093-5. [DOI] [PubMed] [Google Scholar]
- Li X., Krasnyanski S.F., Korban S.S. Optimization of the uidA gene transfer into somatic embryos of rose via Agrobacterium tumefaciens. Plant Physiol. Biochem. 2002;40:453–459. [Google Scholar]
- Linde M., Hattendorf A., Kaufmann H., Debener T. Powdery mildew resistance in roses: QTL mapping in different environments using selective genotyping. Theor. Appl. Genet. 2006;113:1081–1092. doi: 10.1007/s00122-006-0367-2. [DOI] [PubMed] [Google Scholar]
- Lister R., Gregory B.D., Ecker J.R. Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond. Curr. Opin. Plant Biol. 2009;12:107–118. doi: 10.1016/j.pbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Marquardt S., Lister C., Swiezewski S., Dean C. Targeted 3' processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327:94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- Longhi S., Giongo L., Buti M. Molecular genetics and genomics of the Rosoideae: state of the art and future perspectives. Hortic. Res. 2014;1:1. doi: 10.1038/hortres.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnard J.-L., Roccia A., Caissard J.-C. Biosynthesis of monoterpene scent compounds in roses. Science. 2015;349:81–83. doi: 10.1126/science.aab0696. [DOI] [PubMed] [Google Scholar]
- Marquardt S., Raitskin O., Wu Z., Liu F., Sun Q., Dean C. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell. 2014;54:156–165. doi: 10.1016/j.molcel.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T.H.E., Hayes B.J., Goddard M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddam H.H., Leus L., De Riek J., Van Huylenbroeck J., Van Bockstaele E. Construction of a genetic linkage map with SSR, AFLP and morphological markers to locate QTLs controlling pathotype-specific powdery mildew resistance in diploid roses. Euphytica. 2012;184:413–427. [Google Scholar]
- Mohamed R., Wang C.T., Ma C. Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J. 2010;62:674–688. doi: 10.1111/j.1365-313X.2010.04185.x. [DOI] [PubMed] [Google Scholar]
- Nair S.K., Wang N., Turuspekov Y. Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc. Natl. Acad. Sci. U. S. A. 2010;107:490–495. doi: 10.1073/pnas.0909097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y., Higuchi Y., Yoshida Y., Hisamatsu T. Environmental responses of the FT/TFL1 gene family and their involvement in flower induction in Fragaria x ananassa. J. Plant Physiol. 2015;177:60–66. doi: 10.1016/j.jplph.2015.01.007. [DOI] [PubMed] [Google Scholar]
- Oosumi T., Gruszewski H.A., Blischak L.A. High-efficiency transformation of the diploid strawberry (Fragaria vesca) for functional genomics. Planta. 2005;223:1219–1230. doi: 10.1007/s00425-005-0170-3. [DOI] [PubMed] [Google Scholar]
- Otagaki S., Ogawa Y., Hibrand-Saint Oyant L. Genotype of FLOWERING LOCUS T homologue contributes to flowering time differences in wild and cultivated roses. Plant Biol. 2015;17:808–815. doi: 10.1111/plb.12299. [DOI] [PubMed] [Google Scholar]
- Pankin A., Campoli C., Dong X. Mapping-by-sequencing identifies HvPHYTOCHROME C as a candidate gene for the early maturity 5 locus modulating the circadian clock and photoperiodic flowering in barley. Genetics. 2014;198:383. doi: 10.1534/genetics.114.165613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati P.K., Rath S.P., Sharma M., Sood A., Ahuja P.S. In vitro propagation of rose—a review. Biotechnol. Adv. 2006;24:94–114. doi: 10.1016/j.biotechadv.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Pei H., Ma N., Chen J. Integrative analysis of miRNA and mRNA profiles in response to ethylene in rose petals during flower opening. PLoS One. 2013;8:e64290. doi: 10.1371/journal.pone.0064290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H., Ma N., Tian J. An NAC transcription factor controls ethylene-regulated cell expansion in flower petals. Plant Physiol. 2013;163:775–791. doi: 10.1104/pp.113.223388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petterle A., Karlberg A., Bhalerao R.P. Daylength mediated control of seasonal growth patterns in perennial trees. Curr. Opin. Plant Biol. 2013;16:301–306. doi: 10.1016/j.pbi.2013.02.006. [DOI] [PubMed] [Google Scholar]
- Randoux M., Daviere J.M., Jeauffre J. RoKSN, a floral repressor, forms protein complexes with RoFD and RoFT to regulate vegetative and reproductive development in rose. New Phytol. 2014;202:161–173. doi: 10.1111/nph.12625. [DOI] [PubMed] [Google Scholar]
- Randoux M., Jeauffre J., Thouroude T. Gibberellins regulate the transcription of the continuous flowering regulator, RoKSN, a rose TFL1 homologue. J. Exp. Bot. 2012;63:6543–6554. doi: 10.1093/jxb/ers310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remay A., Lalanne D., Thouroude T., Le Couviour F., Hibrand-Saint Oyant L., Foucher F. A survey of flowering genes reveals the role of gibberellins in floral control in rose. Theor. Appl. Genet. 2009;119:767–781. doi: 10.1007/s00122-009-1087-1. [DOI] [PubMed] [Google Scholar]
- Richards E.J. Natural epigenetic variation in plant species: a view from the field. Curr. Opin. Plant Biol. 2011;14:1–6. doi: 10.1016/j.pbi.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Roberts A.V., Gladis T., Brumme H. DNA amounts of roses (Rosa L.) and their use in attributing ploidy levels. Plant Cell Rep. 2009;28:61–71. doi: 10.1007/s00299-008-0615-9. [DOI] [PubMed] [Google Scholar]
- Schneeberger K. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 2014;15:662–676. doi: 10.1038/nrg3745. [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Meth. 2009;6:550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- Semeniuk P. Inheritance of recurrent blooming in Rosa-wichuraiana. J. Hered. 1971;62:203. [Google Scholar]
- Shulaev V., Sargent D.J., Crowhurst R.N. The genome of woodland strawberry (Fragaria vesca) Nat. Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Angel A., Howard M., Dean C. Vernalization – a cold-induced epigenetic switch. J. Cell Sci. 2012;125:3723–3731. doi: 10.1242/jcs.084764. [DOI] [PubMed] [Google Scholar]
- Spiller M., Linde M., Hibrand-Saint Oyant L. Towards a unified genetic map for diploid roses. Theor. Appl. Genet. 2011;122:489–500. doi: 10.1007/s00122-010-1463-x. [DOI] [PubMed] [Google Scholar]
- Sun Q., Csorba T., Skourti-Stathaki K., Proudfoot N.J., Dean C. R-Loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science. 2013;340:619–621. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K., Alonso-Blanco C., Lynn J.R. Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J. 1999;20:67–77. doi: 10.1046/j.1365-313x.1999.00577.x. [DOI] [PubMed] [Google Scholar]
- Swiezewski S., Liu F., Magusin A., Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- Tuskan G.A., Difazio S., Jansson S. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Vamosi J.C., Dickinson T.A. Polyploidy and diversification: a phylogenetic investigation in Rosaceae. Int. J. Plant Sci. 2006;167:349–358. [Google Scholar]
- Vergne P., Maene M., Gabant G., Chauvet A., Debener T., Bendahmane M. Somatic embryogenesis and transformation of the diploid Rosa chinensis cv Old Blush. Plant Cell, Tissue Organ Cult. (PCTOC) 2009;100:73–81. [Google Scholar]
- Verhoeven K.J.F., Van Gurp T.P. Transgenerational effects of stress exposure on offspring phenotypes in apomictic dandelion. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vries D.P. Juvenility in hybrid tea-roses. Euphytica. 1976;25:321–328. [Google Scholar]
- Wang J.-W., Park M.Y., Wang L.-J. MiRNA control of vegetative phase change in trees. PLoS Genet. 2011;7:e1002012. doi: 10.1371/journal.pgen.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W., Bao Y., Luo P., Bao M., Ning G. An efficient system to produce transgenic plants via cyclic leave-originated secondary somatic embryogenesis in Rosa rugosa. Acta Physiol. Plant. 2014;36:2013–2023. [Google Scholar]
- Xing W., Wang Z., Wang X.Q., Bao M.Z., Ning G.G. Over-expression of an FT homolog from Prunus mume reduces juvenile phase and induces early flowering in rugosa rose. Sci. Hortic. 2014;172:68–72. [Google Scholar]
- Yan Z., Visser P.B., Hendriks T., Prins T.W., Stam P., Dolstra O. QTL analysis of variation for vigour in rose. Euphytica. 2007;154:53–62. [Google Scholar]
- Yokoya K., Roberts A.V., Mottley J., Lewis R., Brandham P.E. Nuclear DNA amounts in roses. Ann. Bot. 2000;85:557–561. [Google Scholar]
- Yu C., Luo L., Pan H., Guo X., Wan H., Zhang Q. Filling gaps with construction of a genetic linkage map in tetraploid roses. Front. Plant Sci. 2015;5:796. doi: 10.3389/fpls.2014.00796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Lian H., Wang J.W. Plant developmental transitions: the role of microRNAs and sugars. Curr. Opin. Plant Biol. 2015;27:1–7. doi: 10.1016/j.pbi.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Zakizadeh H., Lütken H., Sriskandarajah S., Serek M., Müller R. Transformation of miniature potted rose (Rosa hybrida cv. Linda) with P SAG12 -ipt gene delays leaf senescence and enhances resistance to exogenous ethylene. Plant Cell Rep. 2012;32:195–205. doi: 10.1007/s00299-012-1354-5. [DOI] [PubMed] [Google Scholar]
- Zhou C., Wang J. Molecular mechanisms of flowering control in perennial herbaceous plants. Chin. J. Cell Biol. 2013;35:1073–1076. (in Chinese) [Google Scholar]
- Zhu Q.H., Upadhyaya N.M., Gubler F., Helliwell C.A. Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa) BMC Plant Biol. 2009;9:149. doi: 10.1186/1471-2229-9-149. [DOI] [PMC free article] [PubMed] [Google Scholar]