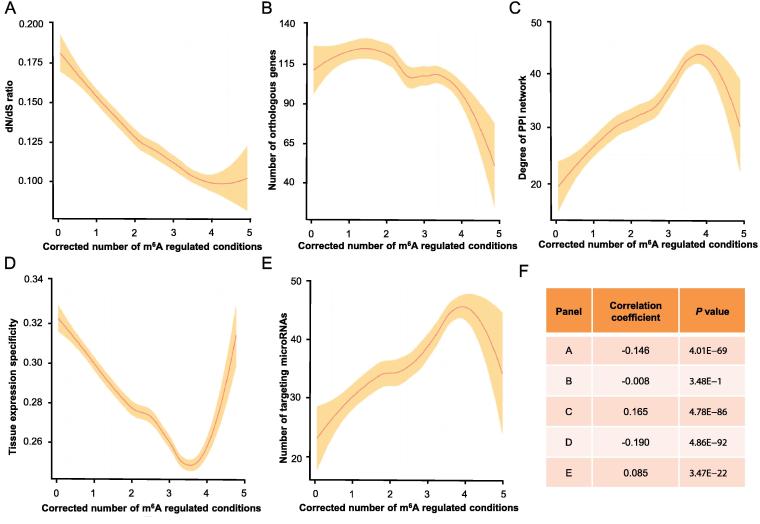
Figure 2.
The correlation between the corrected number of m6A regulated conditions and various gene features
The correlation curves between the corrected number of m6A regulated conditions and various gene features are plotted by using the LOESS smoothing technique. The line indicates the local average estimated by LOESS smoothing and the shade indicates the confidence interval. Outlier genes (0.5%) with extremely high corrected number of m6A regulated conditions are omitted due to their high variation in gene feature values, which could result in badly skewed regression lines. A. Correlation of corrected number of m6A regulated conditions with dN/dS ratio. B. Correlation of corrected number of m6A regulated conditions with number of orthologous genes. C. Correlation of corrected number of m6A regulated conditions with PPI network degree. D. Correlation of corrected number of m6A regulated conditions with tissue expression specificity. E. Correlation of corrected number of m6A regulated conditions with number of targeting microRNAs. F. The summary of Spearman’s correlation coefficient and P values for panels A–E. PPI, protein–protein interaction.