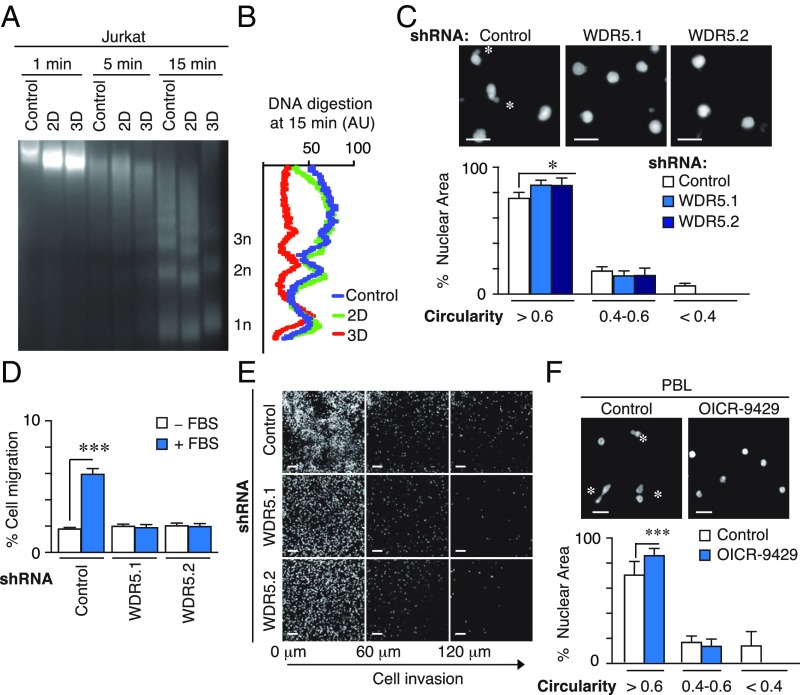
Fig. 4.
WDR5 regulates chromatin conformation and nuclear deformability induced by 3D conditions. (A) Jurkat cells were cultured in suspension, 2D and 3D conditions. Then, cells were digested with micrococcal nuclease (MNase) at indicated times and DNA fragments resolved in agarose gel. (B) Signal profile of the mononucleosomes (1n), dinuclueosomes (2n), and trinucleosomes (3n) after micrococcal digestion (15 min) from cells in A. (C) Control or WDR5-depleted cells were embedded in 3D collagen, fixed and stained with DAPI. Asterisks mark the highly deformable nuclei. Graph shows the circularity index (rounded, >0.6; deformable, 0.4–0.6; or highly deformable, <0.4). Mean n = 3 ± SD. *P < 0.05. (D) Control or WDR5-depleted cells were seeded on collagen gel in the upper chamber of transwells and allowed to invade in response to 10% FBS for 24 h. Cells were collected from the bottom chamber, stained, and quantified. Mean n = 3 ± SD. ***P < 0.001. (E) Cells as in D were fixed after 6 h of cell migration in the collagen gel, stained with DAPI, and serial confocal sections were captured. (F) PBL cells were preincubated or not with OICR-9429, embedded in 3D collagen, fixed and stained with DAPI. Mean n = 3 ± SD. Asterisks mark the highly deformable nuclei. Graph shows the circularity index. Mean n = 3 ± SD. ***P < 0.01.