Abstract
Nuclear receptor subfamily 1, group D, member 1 (Nr1d1) (also known as Rev-erb alpha) has been linked to circadian rhythm regulation, mood-related behavior, and disorders associated with social deficits. Recent work from our laboratory found striking decreases in Nr1d1 in the nucleus accumbens (NAc) in the maternal condition and indirect evidence that Nr1d1 was interacting with numerous addiction and reward-related genes to modulate social reward. In this study, we applied our insights from the maternal state to non-parental adult mice to determine whether decreases in Nr1d1 expression in the NAc via adeno-associated viral (AAV) vectors and short hairpin RNA (shRNA)-mediated gene knockdown were sufficient to modulate social behaviors and mood-related behaviors. Knockdown of Nr1d1 in the NAc enhanced sociability, reduced anxiety, but did not affect depressive-like traits in female mice. In male mice, Nr1d1 knockdown had no significant behavioral effects. Microarray analysis of Nr1d1 knockdown in females identified changes in circadian rhythm and histone deacetylase genes and suggested possible drugs, including histone deacetylase inhibitors, that could mimic actions of Nr1d1 knockdown. Quantitative real-time PCR (qPCR) analysis confirmed expression upregulation of genes period circadian clock 1 (Per1) and period circadian clock 2 (Per2) with Nr1d1 knockdown. Evidence for roles for opioid-related genes opioid receptor, delta 1 (Oprd1) and preproenkephalin (Penk) was also found. Together, these results suggest that Nr1d1 in the NAc modulates sociability and anxiety-related behavior in a sex-specific manner and circadian, histone deacetylase, and opioid-related genes may be involved in the expression of these behavioral phenotypes.
Keywords: Rev-erb alpha, gene expression, AAV vectors, shRNA, gene knockdown
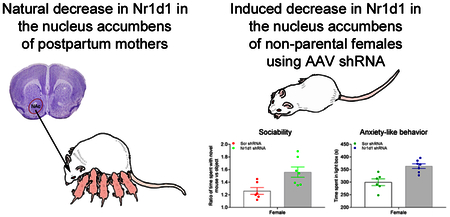
Natural decreases in the circadian gene Nr1d1 in the nucleus accumbens (NAc) of postpartum females likely facilitate reward and prosocial parental behavior. In this study, we found that knockdown of Nr1d1 in the NAc in non-parental female mice enhanced sociability and reduced anxiety-related behavior. We suggest that circadian, histone deacetylase, and opioid-related genes may be involved in the expression of these behavioral phenotypes.
Introduction
Social deficits are strongly coupled to most major mental health disorders, including depression, bipolar disorder (BPD), schizophrenia, and autism (Lewinsohn et al., 1980; Mundy et al., 1986; Hafner et al., 1995; Goldstein et al., 2006). In many cases, social interactions that were once positive and rewarding become aversive and individuals become socially withdrawn. Despite the strong negative impact that social withdrawal and avoidance have on both the affected individual and family, the underlying mechanisms are poorly understood and treatments are lacking. As observed in other mammals, naïve prepartum female mice find social interactions with mouse pups to have low valence (Lonstein & De Vries, 2000), but postpartum females find social interactions with pups to be intensely rewarding (Hauser & Gandelman, 1985). These findings suggest that reward mechanisms that are powerfully altered in association with the initiation of postpartum prosocial behavior may represent fundamental mechanisms that promote sociability across social contexts.
Nuclear receptor subfamily 1, group D, member 1 (Nr1d1) (also known as Rev-erb alpha) is of interest as our recent work found striking and reliable decreases in Nr1d1 in postpartum (i.e., prosocial) compared to prepartum (i.e., asocial) mice across multiple brain regions regulating reward (Eisinger et al., 2013b; Driessen et al., 2014; Eisinger et al., 2014), including the nucleus accumbens (NAc) (Zhao et al., 2014). Nr1d1 is a transcription factor best studied for its role in circadian rhythm regulation (Ueda et al., 2005; Mohawk et al., 2012); however our work and that by others is starting to reveal a key role for Nr1d1 in reward-related processes (Wang et al., 2008; Banerjee et al., 2014; Zhao et al., 2014), mood-related behavior (Kripke et al., 2009; Etain et al., 2011; Chung et al., 2014), as well as other disorders associated with social deficits (Goto et al., 2017). Using large scale gene expression and bioinformatics approaches, evidence from our maternal NAc microarray and qPCR studies suggested that Nr1d1 interacts with and possibly regulates numerous addiction and reward-related genes in the NAc (Zhao et al., 2014). Furthermore, a recent study showed that an Nr1d1 agonist affecting the entire brain inhibited reward (Banerjee et al., 2014). This suggests that decreases in Nr1d1 we observed in the NAc and other brain regions in postpartum females may facilitate reward and prosocial parental behavior. However, to date almost no studies have directly linked Nr1d1 to social behavior or reward. NAc is strongly implicated in reward, including social reward and prosocial behaviors in maternal and non-maternal animals (Insel, 2003). We recently found that almost every neuron in the NAc expresses Nr1d1 (our unpublished observations). Thus, Nr1d1 is a potentially critical gene in linking NAc-mediated reward, social, and mood-related behaviors. What is missing is knowledge of the biological effect of decreases in Nr1d1 in the NAc on social and mood-related behaviors.
In this study, we took novel insights from our work on the postpartum mothers and applied them to non-parental adults. We manipulated Nr1d1 expression in the NAc using shRNA-mediated gene silencing in adult virgin females and males in order to understand the effect of Nr1d1 decreases on reward, social and mood-related behaviors. To identify effects of decreasing Nr1d1 in the NAc on large scale gene expression changes, we used microarray and bioinformatics approaches. Additionally, expression of addiction and reward-related genes that exhibit functional interactions with Nr1d1 in the NAc was assessed using quantitative real-time PCR (qPCR) assay.
Materials and methods
Animals
Mice used in this study were inbred C57BL/6J (stock #000664, The Jackson Laboratory, Bar Harbor, ME, USA) females and males between 10–12 weeks of age at the start of the experiment. All mice were experimentally-naïve when obtained. After arriving at the animal facility, all mice were housed individually until tissue collection. Subjects were provided with ad libitum access to rodent standard chow (Harlan, Madison WI) and water, and were housed with shredded aspen bedding material in polypropylene cages that were changed weekly. All subjects were kept on a 12:12 light/dark cycle with lights on at 6:00 CST. All procedures followed guidelines set by the National Institutes of Health Guide for the Care and use of Laboratory Animals, and were approved by the University of Wisconsin Animal Care and Use Committee. Every effort was made to minimize the number of animals used, and reduce their pain and discomfort.
Short hairpin RNA (shRNA) design and intra-nucleus accumbens injections of AAV vectors
Design, construction and packaging of target adeno-associated viral (AAV) vector (i.e., AAV5-GFP-U6-Nr1d1-shRNA) were carried out by Vector Biolabs (Philadelphia, PA, USA). The shRNA sequences directed toward mouse Nr1d1 were 5′-CCGG-GCGCTTTGCATCGTTGTTCAACTCGAGTTGAACAACGATGCAAAGCGC-TTTTTG-3′. As a negative control, AAV vectors with a scrambled oligonucleotide sequence (i.e., AAV5-GFP-U6-Scr-shRNA, Vector Biolabs, Cat. #7042) with no match with any known genes/mRNAs were chosen. Transcription of the shRNA targeted to the mouse Nr1d1 was driven by a U6 promoter, while Green fluorescent protein (GFP) was co-expressed in the vectors as a reporter gene driven by a CMV promoter. We selected serotype 5 of AAV vectors because AAV5 vectors display higher transduction efficiency in multiple brain regions in different animal models (Davidson et al., 2000; Burger et al., 2004; Markakis et al., 2010). The titers of the packaged viruses for in vivo injections were 1.5×1013 GC/ml. The efficacy of shRNA knockdown of Nr1d1 mRNA was validated in vitro (knockdown of >80% at mRNA level, performed by the vendor). Mice were securely placed on a stereotaxic apparatus (David Kopf Instruments, Tunjunga, CA, USA) and anesthetized with isoflurane at vaporizer setting of 2%. A non-steroidal anti-inflammatory analgesic ketoprofen (Zoetis, Kalamazoo, MI, USA) at a dose of 0.05 mL/10 g body weight sc was administered before a midline anterior to posterior incision was made. Then, local anesthetics with a mixture of bupivacaine/lidocaine 50:50 (Hospira, Lake Forest, IL, USA) were applied on the periosteum. The Bregma was determined and cannula (33 gauge) tip was moved to the coordinates of target site bilaterally according to The Mouse Brain in Stereotaxic Coordinates (Paxinos & Franklin, 2001). AP=1.54 mm, ML=± 0.9 mm, and DV=4.6 mm. A volume of 2.0 uL per side of AAV vectors expressing either mouse Nr1d1-shRNA (i.e., AAV5-GFP-U6-Nr1d1-shRNA) or Scr-shRNA (i.e., AAV5-GFP-U6-Scr-shRNA) were injected bilaterally into the NAc using a Hamilton syringe at a rate of 1.0 uL/min. After injection, the cannula was left in place for additional 5 min. Body weights were monitored and measured once per week following surgery. The number of mice used for microinjection of AAV vectors were as follows: N=26 (13/13 for Nr1d1 shRNA/Scr-shRNA) in females; N=22 (11/11 for Nr1d1 shRNA/Scr-shRNA) in males. To verify the specificity of knockdown of Nr1d1 in the NAc, qPCR analysis was carried out using the reporter gene, GFP. Use of vectors co-expressing GFP enabled us to identify only the neurons transduced with viral-mediated shRNA (i.e. Scr shRNA or Nr1d1 shRNA) and help define the brain region of gene delivery. We performed a pilot qPCR assay to determine GFP expression in the NAc of non-vector-injected mice (n=6) using vendor-provided GFP-specific primers. We found all the Cq values of GFP in each animal (i.e., 32.96, 32.42, 33.19, 32.66, 32.37, 32.30) were above 32. Thus, we adopted a Cq<32 cutoff value to ensure a successful injection and efficient transduction of the viral vectors in the NAc. Only animals injected with viral vectors displayed a Cq value <32 measured in qPCR assay were included for data analysis. We chose the inclusion criteria such that only mice were analyzed that were effectively transduced with the AAV vectors in the NAc neurons.
Social approach test
Behavioral tests were started with the social approach test 3 weeks after injection of AAV vectors, followed by the order of social conditioned place preference, novel object place conditioning, light/dark box test, and forced swim test, with 3- or 4-day intervals between tests. All behavioral testing was conducted during the light phase of the light/dark cycle (i.e., 9:00 AM to 1:00 PM).
Subject mice were Nr1d1- or Scr-shRNA injected C57BL/6J mice and novel mice were C57BL/6J mice between 10 and 12 weeks of age. For pups, pregnant mice were purchased from The Jackson Laboratory and were housed in our animal facility for parturition. Pups at postnatal days 3–6 were used in this test. The apparatus for social approach test is a three-chambered cage with a grey metal floor and two sliding doors (dimension of 5 × 8 cm each) on the two side chambers opening on the central chamber to allow the animal to move freely from one chamber to another (Ugo Basile, Varese, Italy). Each clear Perspex chamber measures 40 × 20 × 22 (length x width x height) cm. The standard grid enclosure is 16 cm tall and has an internal diameter of 7 cm. Each enclosure has a top and bottom of grey PVC and grid bars with a diameter of 3 mm and being spaced 7 mm.
Procedures for social approach test were performed as described in previous work (Silverman et al., 2010; Yang et al., 2011). In advance of experiments, novel mice were trained to habituate to the grid enclosures in the chamber. For each training, the novel mouse was placed in a clean grid enclosure into one side chamber for a total of two 15-min sessions. On testing day, subject mice were brought to the test room and allowed to rest for at least 30 min to acclimate to the room. With the two sliding doors closed, a subject mouse was placed in the center chamber of an entirely empty apparatus for 10 min. After acclimation, by opening both sliding doors, subject mice were allowed to move throughout the chambers freely and behaviors were video recorded by using video capture software for 10 min. A lack of side-preference (i.e., that subjects spend approximately the same amount of time in the two side chambers) was confirmed before proceeding with test for sociability. To start sociability test, the subject mouse was confined to the center chamber before the novel object and the novel mouse were introduced to the side chambers by shutting both sliding doors. A clean empty grid enclosure serving as the novel object was first placed in the middle of one side chamber, between the sliding door and the side wall. Then, another clean grid enclosure holding the novel mouse was placed on the other side chamber. Immediately opening both sliding doors, video recording of mouse behaviors was started. After each subject from a given cage had been tested, it was returned to its original home cage. The test apparatus was cleaned thoroughly with 70% ethanol and tap water between subjects and at the end of each test day. Measures were taken of time spent, time sniffed and number of entries into chamber with novel mouse or with novel object by a single trained observer unaware of the identity of subject mouse. The ratio of time spent with the novel mouse/pups versus object (i.e., time spent with novel mouse/pups divided by time spent with novel object) and the percentage of time spent with novel mouse/pups [i.e., time spent with novel mouse/pups divided by (time spent with novel mouse/pups + time spent with novel object) x 100%] were calculated.
Social conditioned place preference (Social CPP)
Three-compartment social CPP apparatus was divided into three equally sized compartments with two sliding doors (5 × 8 cm) at the base of each delimiting wall between the center and side compartments. The side compartment contained one type of novel bedding (either alpha-dri or paperchip, Shepherd Specialty Papers, Watertown, TN, USA) and the center compartment contained no bedding. We followed a protocol for social CPP described by Dolen et al. (Dolen et al., 2013) for two days of conditioning. A pre-conditioning trial (30 min) was used to establish baseline preference for the two sets of bedding cues. The mouse was placed into the center compartment of social test apparatus, with the two sliding doors open. The amount of time spent freely exploring each compartment was recorded during a 30-min test session. Social conditioning (24 hrs): after pre-conditioning trial, mice were assigned to receive social conditioning (i.e. housed with a non-test normal mouse in their home cage) for 24 hrs on one type of bedding. Isolate conditioning (24 hrs): followed by co-housing, mice were left on the isolate bedding cue (housed without a non-test normal mouse in their home cage) on the other type of bedding for 24 hrs. Post-conditioning trial (30 min): after isolate conditioning, animals received a 30-min post-conditioning trial to establish preference for the two conditioned cues. To this end, the side compartment was lined with either socially-paired or isolation-paired beddings (i.e., alpha-dri or paperchip bedding), respectively. The mouse was placed in the center compartment of social test apparatus with both sliding doors opened to allow the mouse freely explore the entire testing apparatus, and the video recording was started. Bedding assignments (social versus isolate) were counterbalanced for an unbiased design. Preference score (i.e., time spent in side compartment lined with socially-paired bedding minus the time spent in side compartment containing isolation-paired bedding), and normalized social preference score (i.e., time spent in side compartment containing socially-paired bedding, post-conditioning trial divided by pre-conditioning trial) were calculated for comparisons between the experimental conditions.
Novel object place conditioning
We adopted an experimental procedure of the novel object place conditioning that has been previously reported (Bevins & Bardo, 1999; Douglas et al., 2003) with slight modifications. One side compartment had black walls and a paperchip-bedding floor, while the other side compartment had white walls and an alpha-dri-bedding floor. Novel objects used for conditioning contained wood blocks, white PVC pipes, and balls of tin foil. Conditioning trial (days 1–3): one experimental animal was placed into one side compartment in which a novel object located in the back corner was introduced daily (i.e., paired side) for 15 min, while the two sliding doors were shut to confine the mouse to the assigned compartment. The mice always received the novel object in the paired side and the novel object was changed daily. At the end of the initial 15 min session, animal was returned to its home cage. After 1 hr, animal was placed into the other side compartment in which no novel object was introduced daily (i.e., unpaired side) for 15 min. Then, the animal was placed back to its home cage. We counterbalanced order of exposure to the paired side and unpaired side, order in which the novel objects were provided to the animals across days, and the side of the compartment (black versus white) paired with the novel objects. Test day was 24 hrs after the last conditioning trial, the subject mouse was placed in the center compartment of the chamber and no objects were placed in the chamber. Mouse behavior was monitored and recorded for 10 min. Time spent in paired side chamber and percentage of time spent with novel object [time spent in paired side chamber divided by (time spent in paired side chamber + time spent in unpaired side chamber) x 100%] were calculated.
Light/dark box test
We followed our published protocol for light/dark box test (Scotti et al., 2011). The apparatus of light/dark box consists of two compartments that are same in size (24.5 × 14.5 × 25.5 cm) but different in color. One compartment is dark black and has a plastic ceiling (i.e., black portion), and the other is brightly illuminated and open (i.e., light portion). The two compartments were connected by a restricted door (3.8 × 3.8 cm) that is located in the separator wall between the two compartments. Animals were habituated to the experimental room for 30 min before the beginning of the test. The mouse was initially placed in the middle of the dark compartment of the apparatus and allowed to freely move around. The task was then run for 10 min by video recording. An animal was considered to have entered the light or dark compartment if all four paws were placed into that compartment. Following completion of the task, the mice were returned to their home cage. After each trial, all compartments were cleaned with 70% ethanol to prevent a bias based on olfactory cues. Time spent in light portion, latency to first light portion entry and number of transitions between the light and dark portions were assessed for later analysis.
Forced swim test
The procedure for forced swim test has been described in our previous work (Scotti et al., 2011). Briefly, this test was performed in a transparent cylindrical glass container (23 cm height x 17 cm diameter). Prior to each test, the container was filled half way, which is 11.5 cm from the bottom with room temperature (23–25°C) tap water. The animals were brought into the testing room with no acclimation period. At the start of each test, the mouse was gently picked up by its tail and slowly placed in the water. Then, movements including floating, swimming and escaping behaviors were video recorded for a 6-min session and subsequently analyzed off-line. At the end of each 6-min test, animals were removed from the water and gently dried with paper towels and placed back into their home cage with a heat lamp on for 15 min. Immobility was defined as an absence of any movements except for those required to balance the body and keep the head and nose above the water. Latency to first floating, total number of swim bouts and duration of immobility were scored. The first two min of the test are considered as the habituation phase and only the last four min were included in the statistical analysis as in previous reports (Can et al., 2012).
Tissue collection and RNA extraction
Following 72 hrs of completion of the forced swim test, mice were lightly anesthetized with isoflurane and decapitated between 9:00 and 12:00 CST. Brains from Scr- and Nr1d1-shRNA injected mice were dissected alternating between the two groups. After decapitation, vaginal lavage was performed on female animals to determine estrous state. All females were found to be in estrous or diestrous. Brains were flash frozen in isopentane on dry ice and stored at −80°C until being sectioned on a cryostat (Leica CM1850, Bannockburn, IL, USA). Sections of 200-micron thickness were cut and mounted on gelatin-coated glass slides. NAc from Bregma 1.94 to 1.10 mm was dissected bilaterally using a micropunch technique (Makino et al., 1994) under a dissecting microscope. In addition, at the same anatomical level, adjacent tissue of caudate putamen (CPu) and distant area cerebral cortex (CTX) were also collected bilaterally for verification of specificity of Nr1d1 knockdown in the NAc. Samples were subsequently stored at −80°C until RNA extraction. Total RNA was extracted with the Aurum Total RNA Fatty and Fibrous Tissue Kit (Bio-Rad, Hercules, CA, USA) as described in our previous work (Zhao et al., 2012b). The integrity, purity and concentration of RNA were measured as reported. Purified total RNA was stored at −80°C until processed.
High-density oligonucleotide array hybridization
Microarray analysis was performed with the GeneChip Mouse Gene 2.0 ST array (Thermo Fisher Scientific Inc, CA, USA) using targets derived from total RNA isolated from NAc as described above. cDNA for array hybridization was synthesized from 150 ng of total RNA from each single mouse using a GeneChip WT Plus Kit (Thermo Fisher Scientific Inc, CA, USA) according to the manufacturer’s specifications. Briefly, total RNA was used to synthesize double-stranded cDNA, which was then used as a template for anti-sense RNA (cRNA) synthesis. This cRNA was in turn used as a template for a second round of single-stranded cDNA (sense strand cDNA, ss-cDNA) synthesis, and the resultant DNA-RNA hybrids were then treated with RNase H to degrade RNA. The ss-cDNA was then fragmented and biotinylated using GeneChip WT Terminal Labeling Kit (Thermo Fisher Scientific Inc, CA, USA) according to the manufacturer’s specifications. Fragmented and labeled ss-cDNA targets were hybridized with the arrays at 45°C for 16 hours. The hybridized arrays were then washed and stained according to manufacturer specifications, and arrays were scanned on an Affymetrix GC3000 G7 Scanner. Data were extracted and processed from scans using Affymetrix Command Console v. 3.1.1.1229. cDNA synthesis, fragmentation, labeling, array hybridization, staining, and scanning were performed by the University of Chicago Genomics Core.
Probeset level summarization and microarray statistical analysis
Inferential statistics for differential expression between control and knockdown samples were calculated using Affymetrix ® Transcriptome Analysis Console (TAC) software (Barresi et al., 2016) and the Robust Multi-array Average (RMA) approach for data normalization (Irizarry et al., 2003; Dixon et al., 2007). The nominal RMA p-value of 0.03 was used for most analysis. For microarray analysis, a total of 12 (6 Scr-shRNA /6 Nr1d1 shRNA) mice in both females and males were used. These animals were randomly selected from the mice with efficient transduction based on the inclusion criteria (N=6 for Scr shRNA group and N=7 for Nr1d1 shRNA group in female; N=6 for Scr shRNA group and N=6 for Nr1d1 shRNA group in male). One female control injected with Scr shRNA was excluded from microarray data analysis for three reasons: 1) GFP levels were four times higher than any other mouse, leaving open the possibility of non-specific effects of high incorporation of AAV; 2) the weight was more than 2 SD higher than for all other female mice throughout the study, suggesting possible gene expression difference due to weight; and 3) Principal component analysis (PCA) of microarray indicated the individual was an outlier from both groups. The microarray data discussed in this publication, both raw and summarized, have been deposited in NCBI’s Gene Expression Omnibus (GEO), and are accessible through GEO Series accession number GSE110363 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).
Enrichment and drug repurposing analysis
Microarray targets with p-values less than 0.03 were used for both ToppCluster (Kaimal et al., 2010) and NIH DAVID analysis (Huang da et al., 2009). Per1 was included in this analysis as its array p-value was just above 0.03 and its expression change was confirmed via qPCR. The default statistical p-value analysis was used for both. While some databases are shared between the two, each also provides unique analysis. Modular Single-set Enrichment Test (MSET) (Eisinger et al., 2013a) was used to test significant microarray results for enrichment of gene lists with multiple databases. We identified genes that are found in multiple databases for depression, BPD, autism, and schizophrenia, and for transcriptional regulation using identical approaches as in our analysis of microarray data from the postpartum NAc (Zhao et al., 2014). We additionally used MSET to identify genes that are associated with circadian rhythms using the MeSH database (Zhao et al., 2014). We also identified genes that were altered in the NAc in the postpartum condition in our previous (Zhao et al., 2014) and that are among a list of potential top 700 maternal genes that show reliable expression changes in the postpartum state across the CNS (Gammie et al., 2016).
We separately uploaded the up and downregulated genes into NIH LINCS L1000 and used the mimic feature to identify possible drugs with similar actions (Lee et al., 2016; El-Hachem et al., 2017).
Weighted Gene Co-expression Network Analysis (WGCNA) analysis
WGCNA is a system biology approach that has been widely utilized in constructing gene co-expression networks and identifying gene modules in multiple species (Langfelder & Horvath, 2008; Li et al., 2018). In this study, WGCNA was used to identify modules of genes whose expression changes are highly correlated to one another. R software was used for all WGCNA analysis (Zhang & Horvath, 2005; Langfelder & Horvath, 2008) of the top 2000 genes. To generate a weighted network of genes (nodes) and their expression correlations (edges), correlations were raised to a soft thresholding power β, chosen such that the network begins to approach a scale-free topology. Using unsupervised hierarchical clustering, a minimum module size of 30 genes, and a threshold setting for merging modules of 0.25, WGCNA identified seven modules. The modules were exported as a Cytoscape network file, which was manually trimmed to consist of genes of interest and their gene-to-gene correlations. Genes from two modules were visualized with Cytoscape v3.0.
Quantitative real-time PCR (qPCR)
We followed the protocol in our published work (Zhao et al., 2012a; Zhao et al., 2012b). Briefly, a SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA, USA) was used to reverse transcribe 100 ng of RNA to cDNA. The cDNA was then amplified using a SsoFast EvaGreen Supermix kit in a BioRad CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Each sample was run in triplicate and standard amplification procedures were used. Primer sequences for genes of interest and reference genes are shown in Supplementary Table 1. Following amplification, a standard curve was generated to assess the empirical PCR reaction efficiency, and a dissociation curve analysis was performed to insure specificity of PCR products. The expression ratio of mRNA of genes in Nr1d1-shRNA relative to Scr-shRNA (normalized to the reference genes Ywhaz and β-actin) was calculated using a relative expression software tool REST 2009 (Pfaffl et al., 2002). To yield individual relative expression level values for genes Nr1d1 and GFP, mean Ct values obtained from qPCR were transformed according to the Pfaffl method (Pfaffl, 2001; Cordes et al., 2015). The resulting values represent Nr1d1 or GFP mRNA expression normalized against two reference genes Ywhaz and β-actin. These relative expression values were used for statistical analysis of gene expression.
Statistical analysis
Statistical analyses were performed using SPSS 22.0 software (SPSS Inc., Chicago, IL, USA). All values were expressed as mean±SEM. Two-group comparisons in behavioral assays and gene expression were analyzed using unpaired student’s t-test, while within-group comparisons were assessed using paired student’s t-test. Tests for females and males were separated three months and aspects of the testing differed, so analysis of females and males as factors in the same model was not performed. For ensuring an unbiased observation of various behaviors, behaviors were scored by a trained experimenter blind to the group assignments and the intra-rater reliability was 95% or greater. For all statistical analyses of qPCR, body weight and various behaviors, N=6 for Scr shRNA group and N=7 for Nr1d1 shRNA group in female; N=6 for Scr shRNA group and N=6 for Nr1d1 shRNA group in male. P<0.05 was considered significant.
Results
Verification of the specificity and efficacy of Nr1d1 knockdown in the NAc
In our preliminary study, fluorescent immunolabeling of AAV vector-injected brain sections with GFP antibodies showed robust GFP expression in the NAc, with minimum or no spread to the adjacent and distant areas (Fig. S2), indicating that delivery of the AAV vectors containing Scr shRNA or Nr1d1 shRNA (i.e., injection site) was specific to the NAc. However, a successful injection does not guarantee an efficient transduction of shRNA into the NAc neurons. We, furthermore, performed quantitative analysis of mRNA levels of GFP by qPCR and found that mice injected with GFP-shRNA vectors expressed significantly elevated GFP mRNA in the NAc when compared with non-vector-injected animals, suggestive of efficient transduction. Further, no GFP mRNA was found in either the adjacent area, CPu, or the distant area, CTX (Fig. S3). These results demonstrated that knockdown of Nr1d1 was specific to the NAc. By using qPCR data of GFP gene expression to ensure sufficient levels of transduction of AAV in control and experimental groups, the final levels of GFP were almost identical between groups in females (Scr shRNA=2.74±0.85; Nr1d1 shRNA=2.70±0.42) and in males (Scr shRNA=1.10±0.22; Nr1d1 shRNA=1.19±0.27), who were selected for gene and behavioral data analysis.
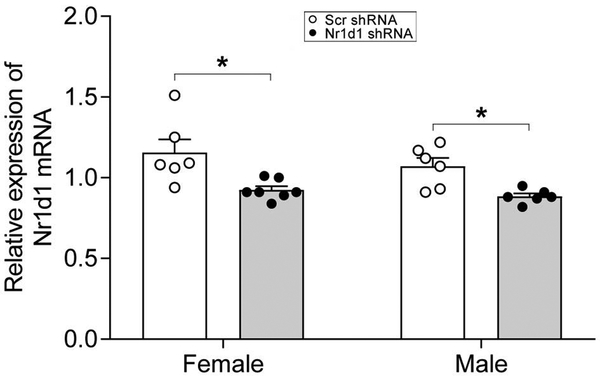
As expected, qPCR data identified that mRNA level of Nr1d1 in the NAc was significantly downregulated in Nr1d1 shRNA-injected group of both females [t(11)=2.92, p=0.036] (by 18.1%) and males [t(10)=3.39, p=0.014] (by 16.8%) as compared to Scr shRNA-injected control group (Fig. 1). To further assess the magnitude of the observed differences between groups, the effect sizes were calculated according to the Cohen’s term d. The value of Cohen’s d was 1.56 for female and 1.95 for male. These decreases are similar to the levels of decreased Nr1d1 in the postpartum NAc (Zhao et al., 2014).
Figure 1:
Combined bar chart and scatter plot of quantitative real-time PCR (qPCR) analysis of expression changes of Nr1d1 in the NAc of shRNA-injected mice. * P < 0.05, Scr-shRNA group versus Nr1d1 shRNA group. NAc, nucleus accumbens; Nr1d1, nuclear receptor subfamily 1, group D, member 1; Scr-shRNA, scrambled short hairpin RNA.
Knockdown of Nr1d1 in the NAc and body weight
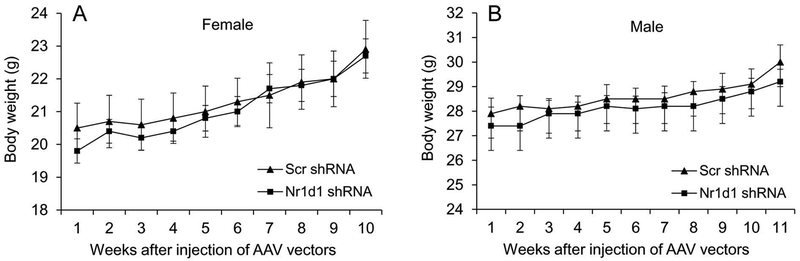
Weight was monitored throughout the study to evaluate indirectly whether changes in feeding occurred with Nr1d1 knockdown. Weight gain occurred in Scr- and Nr1d1-shRNA-injected animals of both females (Fig. 2A) and males (Fig. 2B) over time, but no differences were found between the experimental and control groups.
Figure 2:
Changes in body weight of AAV vector-injected female (A) and male (B) mice. Body weight was measured once per week following vector injection and the average weight of each group was shown. Note that there was no difference in body weight between the Scr- and Nr1d1-shRNA-injected groups in both females and males throughout the study. AAV, adeno-associated virus; Nr1d1, nuclear receptor subfamily 1, group D, member 1; shRNA, short hairpin RNA.
Knockdown of Nr1d1 in the NAc enhanced sociability in female mice
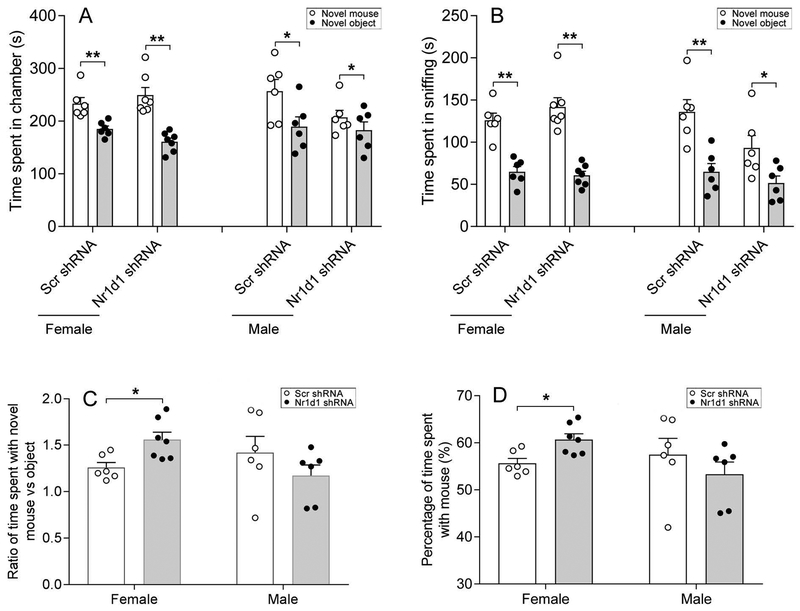
Social behaviors were evaluated using multiple measures. Both females and males injected with Scr shRNA and Nr1d1 shRNA displayed sociability as all groups spent more time in the chamber containing the conspecific (same sex) novel mouse than in a chamber containing the non-social novel object [Female: t(5)=4.83, p=0.005 for Scr shRNA; t(6)=7.03, p<0.001 for Nr1d1 shRNA. Male: t(5)=2.09, p=0.041 for Scr shRNA; t(5)=1.22, p=0.028 for Nr1d1 shRNA] (Fig. 3A) and spent more time sniffing the novel mouse than the novel object [Female: t(5)=7.08, p=0.001 for Scr shRNA; t(6)=9.05, p<0.001 for Nr1d1 shRNA. Male: t(5)=6.48, p=0.001 for Scr shRNA; t(5)=2.42, p=0.04 for Nr1d1 shRNA] (Fig. 3B). The number of entries to side chambers did not differ between the two groups in males or females (data not shown), indicating that knockdown of Nr1d1 had little effect on general exploratory locomotor activity. However, when comparing the ratio of time spent with the novel mouse versus object between the Scr shRNA and Nr1d1 shRNA-injected mice, it was found that female Nr1d1 shRNA-injected mice displayed enhanced sociability, as they spent a greater degree of time with novel mouse relative to novel object as compared to the Scr shRNA mice [t(11)=−2.93, p=0.014] with effect size being 1.67 of Cohen’s d (Fig. 3C). Further, the enhanced sociability in females only was supported by the increased percentage of time spent with novel mouse object [t(11)=−3.08, p=0.01] with effect size being 1.73 of Cohen’s d (Fig. 3D). A similar enhancement of sociability with Nr1d1 knockdown was not found in male mice (Fig. 3C,D).
Figure 3:
Combined bar chart and scatter plot of effects of Nr1d1 knockdown on sociability. Sociability was assessed using social approach test with the conspecific mouse serving as the novel target mouse in both female and male mice. Time spent in chamber (A), time spent in sniffing (B), ratio of time spent with novel mouse versus object (C), and percentage of time spent with novel mouse (D). * P < 0.05, ** P < 0.01, novel mouse versus novel object in A and B; * P < 0.05, Scr-shRNA group versus Nr1d1 shRNA group in C and D.
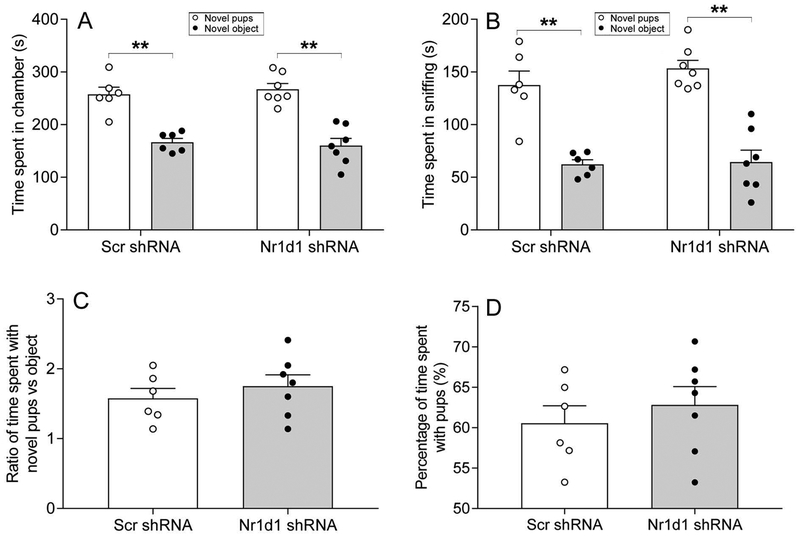
A second test for measuring sociability was also run in female mice, but using pups instead of conspecifics. Like the conspecific mouse, both Scr shRNA and Nr1d1 shRNA-injected mice displayed sociability, as reflected by more time spent in chamber containing the conspecific novel pups than in chamber containing the non-social novel object [t(5)=4.67, p=0.005 for Scr shRNA; t(6)=6.12, p=0.001 for Nr1d1 shRNA] (Fig. 4A), and more time spent sniffing the novel pups than the novel object [t(5)=5.31, p=0.003 for Scr shRNA; t(6)=8.51, p<0.001 for Nr1d1 shRNA] (Fig. 4B). There was no difference in the number of entries to the side chambers between the two groups of mice (data not shown). When comparing the ratio of time spent with novel pups versus object as well as the percentage of time spent with pups between the Scr shRNA and Nr1d1 shRNA-injected mice, Nr1d1 knockdown mice tended to spend more time with pups, but this did not reach statistical significance [t(11)=−0.79, p=0.148 for ratio; t(11)=−0.71, p=0.49 for percentage] (Fig. 4C,D).
Figure 4:
Combined bar chart and scatter plot of effects of Nr1d1 knockdown on sociability. Sociability was assessed using social approach test with pups serving as the novel target mouse in female mice. Time spent in chamber (A), time spent in sniffing (B), ratio of time spent with novel pups versus object (C), and percentage of time spent with novel pups (D). ** P < 0.01, novel pups versus novel object in A and B.
A third test, Social CPP, was used to measure social reward-related behavior in female mice. However, Nr1d1 knockdown mice had a higher pre-conditioning preference score in the pre-conditioning trial (785.8±82.9 sec) relative to control (423.8±38.4 sec). Although the post-conditioning preference score was still higher for Nr1d1 knockdown (598.8±47.7 sec) relative to control (517.6±144.7 sec), no significant differences were found in terms of normalized preference score (1.09±0.14 sec for control; 0.86±0.05 sec for Nr1d1 knockdown).
Knockdown of Nr1d1in the NAc had no effect on general reward-related behavior in both female and male mice
In order to evaluate whether general reward-related (i.e., novelty reward in this study) processes were influenced by decreased Nr1d1 in the NAc, we carried out novel object place conditioning test. No effect of Nr1d1 knockdown was found in time spent in paired side chamber (female: 163.2±13.4 sec for control; 164±7.8 sec for Nr1d1 knockdown; male: 250.7±49.4 sec for control, 200.7±39.5 sec for Nr1d1 knockdown) and percentage of time spent with novel object (female: 44.1±3.9 for control, 43.3±2.0 for Nr1d1 knockdown; male: 52.2±10.0 for control, 45.1±10.0 for Nr1d1 knockdown) for either females or males.
Knockdown of Nr1d in the NAc resulted in anti-anxiety effects in female mice
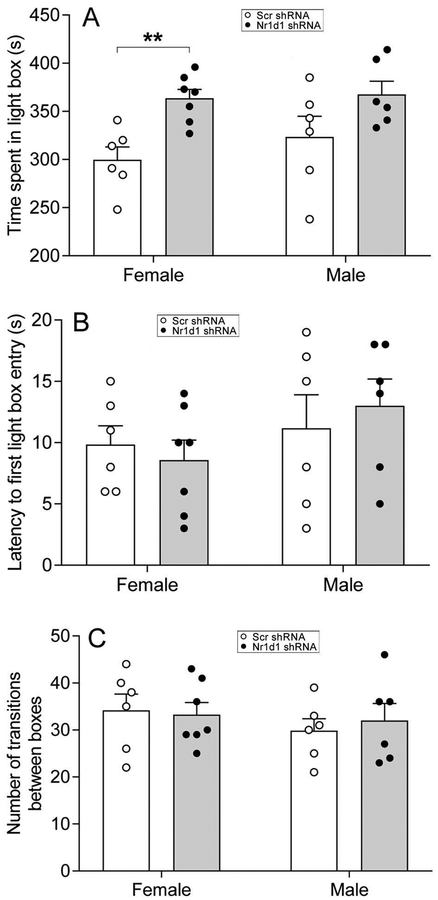
To determine whether Nr1d1 knockdown in the NAc would influence anxiety-related behavior, we performed light/dark box test. In females, Nr1d1 shRNA-injected mice spent significantly more time in light box than Scr shRNA-injected mice [t(11)=−4.02, p=0.002] with effect size being 2.21 of Cohen’s d (Fig. 5A), while no differences in latency to first enter the light box and the number of transitions between boxes were observed (Fig. 5B,C). Similar to females, male mice injected with Nr1d1 shRNA exhibited a trend towards an increase in the amount of time spent in light box when compared with the male mice injected with Scr shRNA, although differences did not reach statistical significance [t(11)=−1.74, p=0.11] (Fig. 5A). Neither the latency to first enter the light box nor the number of transitions between boxes differed between the two groups in male mice (Fig. 5B,C).
Figure 5:
Combined bar chart and scatter plot of effects of Nr1d1 knockdown on anxiety-related behavior measured by light/dark box test. Time spent in light box (A), latency to first enter the light box (B) and number of transitions between boxes (C). ** P < 0.01, Scr-shRNA group versus Nr1d1 shRNA group.
Knockdown of Nr1d1 in the NAc did not influence depression-like behavior
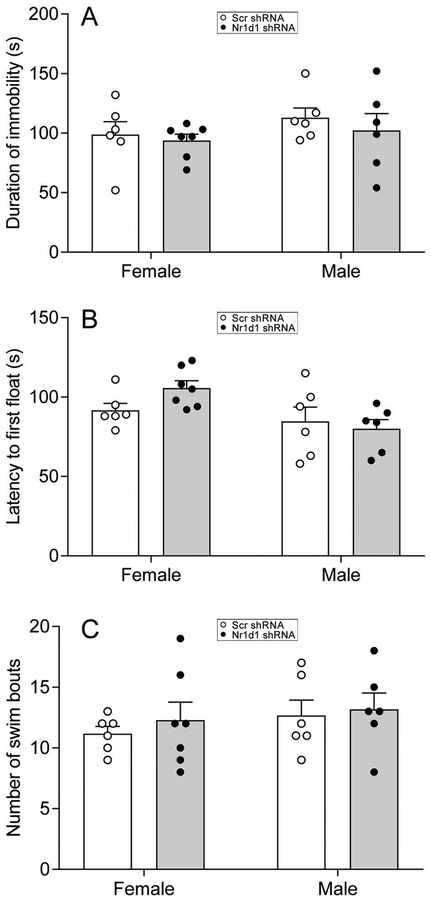
To assess whether decreasing Nr1d1 in the NAc would alter depression-like behavior, we conducted the forced swim test. All the parameters measured, including duration of immobility (Fig. 6A), latency to first float (Fig. 6B) and total number of swim bouts (Fig. 6C) were similar between Scr- and Nr1d1-shRNA-injected mice in both female and male mice.
Figure 6:
Combined bar chart and scatter plot of effects of Nr1d1 knockdown on depression-like behavior examined by forced swim test. Duration of immobility (A), latency to first float (B) and number of swim bouts (C). Note that knockdown of Nr1d1 had no significant effects on depression-like behavior.
Microarray gene expression analysis of NAc with knockdown of Nr1d1 in females and males
Using microarrays, altered gene expression was found with Nr1d1 knockdown in the NAc for 1202 unique, annotated genes in females and for 620 unique, annotated genes in males using a p<0.03 cutoff. Given the lack of behavioral effects of knockdown in males and that the number of significant genes in males was much lower than in females, subsequent analysis reported below focusses on results in females. The full set of results for both females and males is available in Supplementary Table 2. Microarray results, including CEL files, have been uploaded to NCBI’s Gene Expression Omnibus (Accession number: GSE110363).
Pathway analysis in females
NIH DAVID enrichment analysis identified acetylation and mitochondrion as categories with the highest enrichment with Nr1d1 knockdown in females. A list of these genes is provided in Supplementary Table 3. Further, NIH DAVID identified an involvement of histone deacetylation related genes, including Hdac3, Hdac4, Hdac6, Morf4l1, and Suds3. ToppCluster similarly found connections to acetylation and mitochondrion. Both NIH DAVID and ToppCluster found enrichment for Parkinson’s and Huntington’s disease and the list of those genes are provided in Supplementary Table 3.
We additionally used MSET to identify overlap of Nr1d1 knockdown genes in females with various datasets, including addiction, where genes of interest included: Gabra2, Grin2b, Zcrb1, Frmd4a, Oprd1, Per1, and Per2. We evaluated three mental health disorders that can include social deficits, namely depression, bipolar disorder (BPD), and autism and among the genes of interest were: Gabra2, Bcl2, Grin2b, Per1, Per2, Bcl2, Oprd1, Nfil3, and Cacna1h. 87 of the top regulated genes are involved in transcriptional regulation and some of the genes are: Morf4l1, Hdac3, Hdac4, Hdac6, Foxn2, Per1, Per2, and Hif1a. Among circadian rhythm genes were: Per1, Per2, Hdac3, Nfil3, Hnrnpu, Ncoa3, and Cirbp. Additionally, when evaluating a list of potential top 700 maternal genes, overlapping genes of interest included: Morf4l1, Hdac4, Cirbp, Ethe1, and Hif1a. When considering top genes altered in the NAc during the postpartum period, overlapping genes of interest included: Morf4l1, Per1, Per2, Hdac4, Cirbp, Ethe1, and Hif1a. In terms of overlap with the top male genes due to Nr1d1 knockdown (p<0.04 cutoff for both), two genes of interest were: Phf7 and Trim28. Findings are presented in Supplementary Table 3.
L1000CDS analysis
Using NIH LINCS tool, L1000CDS, that allows for analysis of up versus down regulated genes with Nr1d1 knockdown using a p<0.04 cutoff, we found a number of chemicals that were able to mimic some gene aspects of Nr1d1 knockdown. Two of these were parthenolide and vorinostat, both which can act as histone deacetylase inhibitors. Also identified were noretynodrel, which can act as a progestin, Ly 288513, which is a cholecystokin receptor 2 antagonist with anxiolytic properties, modafinil, which can affect arousal, JZL-184, which affects endocannabinoid signaling, and celastrol, that may restore leptin sensitivity (Liu et al., 2015). Identification of drugs that mimic up and down regulated genes in female Nr1d1 knockdown array is summarized in Supplementary Table 4.
qPCR analysis of a subset of addiction and reward-related genes
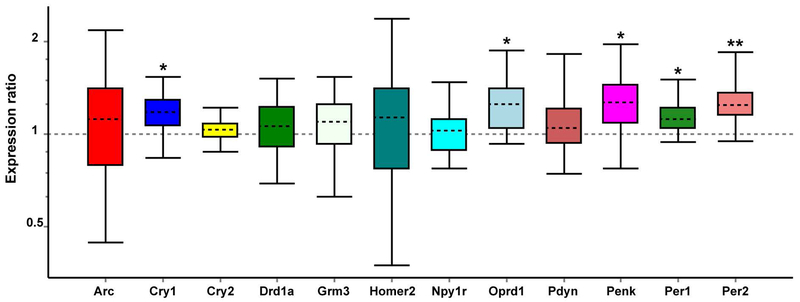
We sought to confirm some of our significantly altered microarray genes and to evaluate other addiction and reward-related genes of interest identified in our recent postpartum NAc study (Zhao et al., 2014) using qPCR assay in females. As shown in Fig. 7, we were able to confirm both Per1 (p=0.018) and Per2 (p=0.007) increases as seen in the microarray. Unexpectedly, we found Oprd1 to be significantly upregulated (p=0.018), which was the opposite pattern as seen in the microarray. When evaluating other circadian and addiction related genes that were altered in the postpartum NAc study (Zhao et al., 2014), but were not among the top altered genes in the present microarray, we found significant increases in expression of Cry1 (p=0.044) and Penk (p=0.037), but a lack of change in other genes, such as Pdyn and Drd1a.
Figure 7:
Quantitative real-time PCR analysis of expression changes for addiction and reward-related genes in the female NAc. Relative expression distribution of mRNA (y-axis) represented as a ratio of Nr1d1 shRNA versus Scr-shRNA, was normalized against two reference genes, Ywhaz and β-actin, and shown by box-and-whisker plot as medians (dashed lines), interquartile range (boxes) and ranges (whiskers). Ratios over one indicate genes with higher expression in Nr1d1 shRNA than in Scr-shRNA mice. * P < 0.05, ** P < 0.01, Nr1d1 shRNA mice versus Scr-shRNA control.
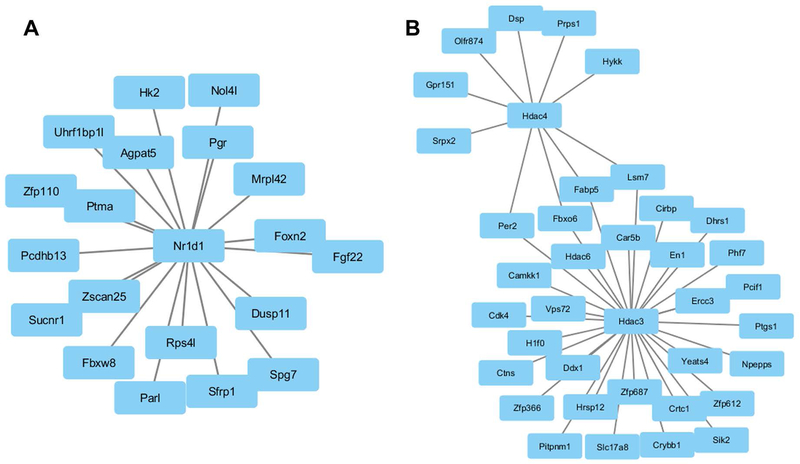
WGCNA analysis
We used WGCNA to identify modules of genes that cluster together based on co-expression. This approach can provide indirect insights into transcriptional regulatory networks. As shown in Fig. 8, we found Nr1d1 expression to be connected to a subset of genes, including the progesterone receptor (Pgr) (Fig. 8A). We found a cluster of genes regulated by both Hdac3 and Hdac4 and a separate group of genes solely regulated by Hdac3 (Fig. 8B).
Figure 8:
Coexpression network analysis using WGCNA. (A) Nr1d1 is connected via coexpression to a subset of genes, including progesterone receptor (Pgr). (B) Two histone deacetylase genes, Hdac3 and Hdac4, each are part of a unique coexpression network, but also have shared genes, including Per2 and Fabp5.
Discussion
This study extended our previous work on social reward and mood state from mothers to non-parental adults. Using a region-specific shRNA-mediated gene silencing technique, we for the first time demonstrated that knockdown of circadian gene Nr1d1 in the NAc potentiated sociability in female mice. Intra-NAc decreases of Nr1d1 also produced an anti-anxiety effect only in females, whereas it had no effects on depression-like behavior in either females or males. Microarray and qPCR analysis demonstrated that intra-NAc knockdown of Nr1d1 altered expression of circadian, histone, and opioid related genes. These findings suggest that Nr1d1 in the NAc modulates sociability and anxiety-related behavior in a sex-specific manner, possibly via circadian, histone, or opioid mediated mechanisms.
Role of Nr1d1 in the NAc on social behavior
Our previous work on NAc found that in the highly social postpartum condition, Nr1d1 expression was significantly decreased and that Nr1d1 may be interacting with a large number of reward/addiction-related genes in the NAc to modulate social reward (Zhao et al., 2014). One striking finding of the present study is that knockdown of Nr1d1 in the NAc enhanced sociability in female adults, but had little effect in males. Thus, the findings support the idea that decreases in Nr1d1 expression contribute to elevated sociability. The upregulation of Per1 and Per2 with Nr1d1 knockdown is consistent with known feedback interactions of these circadian rhythm genes (Hastings et al., 2003) and mirrors our previous finding of elevated Per1 and Per2 in the NAc in the postpartum condition (Zhao et al., 2014). Circadian genes play a critical role in addiction, motivation, and reward (Falcon & McClung, 2009), although a link to sociability is less well studied. Additional circadian genes altered by Nr1d1 knockdown included Nfil3, which is also found in databases for depression and autism, two disorders with social deficits (see Supplementary Table 3).
Pharmacological, molecular and electrophysiological studies have demonstrated an integral role for NAc in reward-related behavior, including dominant roles for the dopamine and opioid signaling pathways (Ikemoto et al., 1997; Carlezon & Thomas, 2009; Trezza et al., 2011). The endogenous opioid neuropeptide, enkephalin (Penk), can act via mu and delta opioid receptors in the NAc to induce rewarding effects (Goeders et al., 1984; Raynor et al., 1994; Simmons & Self, 2009; Trezza et al., 2011). Given that Nr1d1 knockdown altered expression of Penk and Oprd1, it is possible that some modulation of sociability is occurring via these opioid pathways. Further, we have found that a large number of enkephalin neurons co-express Nr1d1 in the NAc (our unpublished observations), providing neuroanatomical evidence for an interaction of Nr1d1 and Penk signaling. The interaction of Nr1d1 with Penk may be complex as in our postpartum NAc study (Zhao et al., 2014), we found indirect evidence that lower Nr1d1 was linked to lower Penk, but here it is connected with higher Penk. We identified additional genes altered by Nr1d1 decreases that are known to be involved addiction and reward pathways (Supplemental Table 2), and these included Gabra2, Grin2b, Zcrb1, and Frmd4a. Additional analysis is needed to understand how these reward-related genes may connect with Nr1d1.
The finding of genes involved in histone deacetylase signaling (Hdac3, Hdac4, Hda6, and Suds3) and that acetylation pathways were enriched by Nr1d1 knockdown (including Yeats4, Eny2, and Morf4l1) suggests Nr1d1 is affecting transcriptional regulation at multiple levels. The WGCNA analysis highlighted how Hdac3 and Hdac4 have converging actions on a subset of genes and that each are strong nodes for expression regulation. Interestingly, one gene regulated by Hdac3 is H1f0 (Fig. 8B). H1f0 was found recently to be a possible hub gene in the NAc that could contribute to elevated motivation to exercise in mice (Saul et al., 2017) and here it could also be regulating changes in sociability. One gene, Morf4l1, is a component of the NuA4 histone acetyltransferase complex (Zou & Mallampalli, 2014) and stands out for a few reasons. In addition to contributing to transcriptional regulation, it also showed altered expression in the NAc in the postpartum condition, it is considered one of the top 700 maternal genes that act in the CNS (Gammie et al., 2016), and it showed altered expression in ventral tegmental area in Nr1d1 knockout mice (Chung et al., 2014). Morf4l1 interacts with Yeats4, which is also a component of the NuA4 complex and is another gene affected by Nr1d1 knockdown. Morf4l1 could be a key downstream target of Nr1d1 that plays an important role in altering social behavior via its transcriptional actions. Using WGCNA, we found an interaction between Nr1d1 and the progesterone receptor, Pgr (Fig. 8A). Given the high importance of progesterone signaling in the development of the maternal brain (Numan et al., 1999; Sheehan & Numan, 2002), Pgr could be a mechanism by which Nr1d1 alters social behavior.
To gain additional insight how Nr1d1 may regulate sociability we looked for genes that are associated with mental health disorders that include social deficits, namely depression, BPD, and autism. In addition to the circadian genes listed above (Per1, Per2, and Nfil3), we identified as genes of interest: Gabra2, Tsc2, Grin2b, and Bcl2 (Supplementary Table 3). We also looked for genes that are either top maternal genes across the CNS (Gammie et al., 2016) or are altered in the NAc in the postpartum condition as either could be related to the emergence of prosocial behavior in mothers. Among these maternal genes are: Morf4l1, Hdac4, Cirbp, Ethe1, and Hif1a (Supplementary Table 3). Whether or how any of these genes contributes to alterations in social behavior is still to be determined.
One unexpected finding was the enrichment of Nr1d1 knockdown genes with datasets for both Huntington’s and Parkinson’s disease (Supplementary Table 3). The enrichment stems in part from the high number of NADH: Ubiquinone Oxidoreductase related genes (n=7) affected by Nr1d1. Whether variation in Nr1d1 expression has any bearing on the typical development of either disorder would need to be addressed in follow up studies.
Given that Nr1d1 knockdown elevated some aspects of sociability, we were interested if there were any drugs that might mimic the effects of Nr1d1 decreases on gene expression and thus be possible candidates for social behavioral disorder treatments. Using NIH LINCS program, we found a subset of drugs that mimicked Nr1d1 knockdown and these included parthenolide and vorinostat, both of which can act as histone deacetylase inhibitors (Gopal et al., 2007; Tiffon et al., 2011). Interestingly, vorinostat and other HDAC inhibitors are being investigated for anti-depressive actions (Meylan et al., 2016; Powell et al., 2017) and infusions into NAc mitigate stress effects and lead to gene expression profiles similar to the anti-depressant, fluoxetine (Covington et al., 2009). Norethylnodrel, a progestin, was also identified and this could be of interest given the finding of Nr1d1 interaction with the progesterone receptor in the WGCNA analysis. LY 288513 (a CCK2 antagonist) can mitigate panic like behavior (Griebel et al., 1997), so the action has some similarities to the anxiolytic effects of Nr1d1 knockdown. The identification of modafinal, the arousal drug, is of interest given how integrated Nr1d1 is in the circadian rhythm pathway regulation. Celastrol is also of interest as it recently has been implicated in restoring leptin sensitivity (Liu et al., 2015).
General reward and depression-like traits were not affected by Nr1d1 knockdown
Feeding involves reward pathways, but as shown in Fig. 2, no effect of Nr1d1 knockdown was found on weight gain in either sex. Further, we did not see an effect of Nr1d1 knockdown on general reward in female or male mice in terms of novel object place conditioning. Sociability can differ from general reward as lactating mothers exhibit increased responses towards their offspring (social reward), but decreased responses to drugs of abuse (general reward) (Mattson et al., 2001; Mattson et al., 2003; Mattson & Morrell, 2005). The finding of alterations in sociability, but not general reward with Nr1d1 knockdown is consistent with the idea that the different rewarding stimuli contain unique components for activation. In this study, we did not see evidence of elevated sociability towards pups, only towards conspecific adult females. One possibility for the lack of response to pups is that we limited exposure, such that the pups were behind a wire mesh. Full interaction with pups may provide different insights into effect of Nr1d1 knockdown and this could be performed in future studies.
When testing for depression-like traits using the forced swim test, no effect of Nr1d1 knockdown was found in either males or females. A growing number of animal studies have demonstrated a role for the NAc in depression-like behavior (Newton et al., 2002; Shirayama & Chaki, 2006; Warner-Schmidt et al., 2012; Francis et al., 2015). Further, abnormalities in circadian rhythm due to perturbations of circadian genes are associated with depression (Kronfeld-Schor & Einat, 2012). For example, increased depression-like behavior is associated with a decreased Per1 and Per2 expression in the NAc, which can be reversed by antidepressant treatment (Spencer et al., 2013). Based on the findings that Nr1d1 knockdown increased Per1, Per2, it might have been expected to see an antidepressant-like effect with Nr1d1 knockdown, but this was not observed. It is possible that modulation of expression of additional genes is required in the NAc to induce a change in this trait.
Role of Nr1d1 in the NAc on anxiety-related behavior
Circadian genes are implicated in the regulation of mood state. Circadian rhythm disturbances evoke a large number of psychiatric disorders such as depression, anxiety, bipolar disorder and autism (McClung, 2007). Nr1d1 is an important circadian gene involved in a wide range of processes, including transcriptional regulation, emotional state, and multiple mental health disorders (Preitner et al., 2002; Solt et al., 2012; Chung et al., 2014; Everett & Lazar, 2014). NAc is also involved in anxiety-related behavior (Zarrindast et al., 2008). In this study we found that knockdown of Nr1d1 in the NAc led to an anxiolytic effect as well as an increase in Per1 and Per2 gene expression in female mice. Our findings are consistent with previous research found that knockout of both Per1 and Per2 enhanced anxiety-related behavior, and that knockdown of Per1 and Per2 specifically in the NAc elevated anxiety (Spencer et al., 2013). Activation of mu- or delta-opioid receptors are anxiolytic (Saitoh et al., 2004; Zarrindast et al., 2008) and alterations in opioid signaling from Nr1d1 knockdown could also contribute to anti-anxiety actions. Interestingly, whole body and CNS exposure to an Nr1d1 agonist can be anxiolytic (Banerjee et al., 2014) and one explanation is that Nr1d1 has non-linear effects on anxiety that are regions specific. Multiple genes downstream from Nr1d1 could underlie the changes in anxiety, but this would need to be evaluated in future work.
Sex differences in response to Nr1d1 knockdown
As noted above, the behavioral effects of Nr1d1 knockdown were specific to females. Further, the gene expression changes in males were fewer than in females, although there were some similar changes with Nr1d1 knockdown. The female-specific effect of Nr1d1 on sociability in non-parental adults is consistent with our findings in postpartum mothers. As Nr1d1 expression is regulated by estradiol (Shimizu et al., 2010), steroid hormones may be a regulator involved in this sex-specific effect of Nr1d1. Converging human and animal studies have revealed sex differences in anxiety, with the prevalence of anxiety disorders being higher in females than in males (Lewinsohn et al., 1998; Piccinelli & Wilkinson, 2000). Circadian genes such as Clock and Per2 are involved in female reproduction and estrous cycle and sex hormones in turn influence circadian rhythm (Miller et al., 2004). Sex differences in gene expression in the CNS have been identified (Rinn & Snyder, 2005; Trabzuni et al., 2013) and it is possible that on this background of different preexisting levels of gene expression that modulation of Nr1d1 is less effective in males than in females. Thus, it is likely that sex hormones contribute to the sex difference in anxiety-related behavior after Nr1d1 knockdown in a complicated manner.
Technical considerations and limitations
Differing from the gene delivery in vitro, many factors can influence the efficiency of gene silencing in vivo. These include types of cell, tissue, vector, serotypes and titers of viral vector (Mason et al., 2010; de Solis et al., 2017). An additional contributing factor to efficiency of gene knockdown is that knockdown of a given gene may trigger potential compensatory mechanisms to buffer against the knockdown effect (Pendaries et al., 2014). In this study, we used shRNA-mediated gene silencing, a powerful and widely used method for gene knockdown in the CNS. We evaluated Nr1d1 knockdown throughout the entire NAc by measuring the overall expression level of Nr1d1 mRNA using qPCR. Our results showed about a 20% of gene knockdown, consistent with previous similar studies that detected a 20% (Choi et al., 2012) and 36% knockdown (Hommel et al., 2003). Moreover, in recent published work from our laboratory, we found that a 20% expression change in mRNA is sufficient to translate a corresponding change at protein level (Zhao et al., 2012a), suggesting that changes in Nr1d1 expression are real and biologically significant.
Like other core circadian genes, Nr1d1 is expressed in a circadian pattern in the CNS (Rath et al., 2013; Rath et al., 2014). In addition, Nr1d1 modulates multiple behavioral processes in a daily rhythmic manner (Chung et al., 2014), which is dependent on its “non-clock” function controlled by brain regions outside of the master circadian pacemaker, the hypothalamic suprachiasmatic nucleus (SCN) (Uz et al., 2005). In this study, we were not able to perform a time course (i.e., multiple time points) of behavioral and molecular tests due to time conflict among multiples tests. Future time-course study is needed to provide deep insights into how Nr1d1 regulates numerous behaviors in a rhythmic manner.
Microarray gene expression analysis of NAc with knockdown of Nr1d1 did not show detectable changes in Baml1 and Clock, two core components of circadian clock. As Nr1d1 acting as a key circadian regulator represses Bmal1 transcription (Preitner et al., 2002), it would be expected to see upregulation of Bmal1 after Nr1d1 knockdown. Contrary to our expectation, no change in Bmal1 was found. It is not clear why Nr1d1 knockdown had little effect on Bmal1. Given that changes in other clock genes (e.g., Per and Cry) were not really big (see Fig. 7), it raises questions about whether these changes are an effect of Nr1d1 knockdown or a shift in the circadian cycle. Our results support a knockdown effect on the basis of two reasons. First, Nr1d1 reaches its peak of expression at Zeitgeber time (ZT) 8–9 (Torra et al., 2000; Mollema et al., 2011; Rath et al., 2013), while all the measurements/experiments were conducted just at one time point, and the vast majority of brain tissues for analysis of gene expression were sampled at ZT 3–4, when Nr1d1 expression is low. Thus, the comparison of clock genes including Baml1, Clock, Cry1, Cry2, Per1 and Per2 between the Scr control and Nr1d1 knockdown mice was made when these genes are expressed at high level in non-mutant control mice and constant high level in Nr1d1 knockdown mice. This may explain in part a lack or little effect of Nr1d1 knockdown on expression of these clock genes. Second, knockdown of Nr1d1 in the NAc led to an anxiolytic effect, consistent with previous findings showing that knockout of both Per1 and Per2 enhanced anxiety-related behavior, and that knockdown of Per1 and Per2 specifically in the NAc elevated anxiety (Spencer et al., 2013). Future work is needed to investigate the time course effect of Nr1d1 knockdown on multiple behavioral phenotypes.
Conclusions
Results from this study indicate that induced decreases in expression of the circadian gene, Nr1d1, in the NAc can enhance sociability and reduce anxiety-related behavior. The findings are consistent with earlier work that identified natural decreases in Nr1d1 expression in postpartum mothers that are associated with the emergence of prosocial behaviors. The regulatory role of the transcription factor Nr1d1 is likely achieved by triggering the multiple downstream events, including actions on circadian, histone, and opioid systems. Future studies can examine the complex ways by which Nr1d1 contributes to natural changes in social behavior.
Supplementary Material
Double immunofluorescence labeling of Nr1d1 (A) with NeuN (neuronal marker, B) showing that the vast majority of Nr1d1-expressing cells are neurons in the NAc of mouse brain. Double fluorescence labeling was carried out with a tyramide signal amplification (TSA) method. Sections were incubated overnight at 4°C with mouse anti-Nr1d1 primary antibodies (LifeSpan BioSciences, LS-B4487, diluted 1:2000). After primary antibody incubation, sections were washed and incubated for 1 hr with HRP-conjugated horse anti-mouse antiserum (Cell Signaling Technology; #7076, diluted 1:100). After several washes, they were incubated for 10 min in Cy3-conjugated tyramide (TSATM Plus Cyanine 3 kit, PerkinElmer, Waltham, MA; red for Nr1d1 labeling) by diluting TSA stock solution 1:50 in 1× Amplification Diluent. After washing with wash buffer, sections were incubated in heated citric acid buffer (10 mM, pH 6.0) at 95°C for 5 min to eliminate HRP activity from the initial TSA reaction and cross-reaction between IgGs raised in the mouse. Sections were then incubated overnight at 4°C with mouse anti-NeuN (Millipore, MAB377, diluted 1:000) primary antibodies. Sections were washed and incubated for 1 hr with HRP-conjugated horse anti-mouse antiserum. After 3 washes, they were incubated for 30 min in Alexa Fluor 488-conjugated tyramide (Molecular Probes, Eugene, OR; green for NeuN labeling) by diluting TSA stock solution 1:100 in 1×Amplification reagent. Following washing 3×10 min with wash buffer, sections were mounted onto slides using DePeX mounting medium (Serva, Heidelberg, Germany), air-dried and stored in the dark at 4°C. Note that the vast majority of Nr1d1-expressing cells were colocalized with NeuN (C), suggesting that they are neurons. Arrows indicate the typical examples of double-labeled neurons in the NAc. Scale bar = 50 μm.
Verification of the injection site and extent of viral transduction in mice injected with AAV vectors expressing Nr1d1 shRNA into the NAc. Immunofluorescence labeling with GFP antibody that serves as a reporter to label the transduced neurons was performed. Brain tissue was collected 3 weeks after stereotaxic injection of AAV Nr1d1 shRNA. Sections were incubated overnight at 4°C with rabbit anti-GFP primary antibodies (Molecular Probes, A-11122, diluted 1:1000). After primary antibody incubation, sections were washed and incubated for 2 hrs with Alexa Fluor 488-conjugated donkey anti-rabbit antiserum (Molecular Probes; A-21206, diluted 1:100). Two representative lower (A, B) and one higher (C) magnification photomicrographs show that the GFP-positive cells were predominantly expressed in the NAc, while sporadically distributed in the adjacent areas. Scale bars, 50 μm in A, B; 20 μm in C.
Quantitative real-time PCR analysis of Nr1d1 and GFP gene expression in the CPu and CTX of female and male mice. Relative expression distribution of mRNA (Y-axis) represented as a ratio of Nr1d1 shRNA-injected versus Scr shRNA-injected mice, was normalized against two reference genes Ywhaz and ?-actin, and shown by box-and-whisker plot as medians (dashed lines), interquartile range (boxes) and ranges (whiskers). Note that no significant differences were observed in mRNAs of Nr1d1 and GFP in CPu and CTX between the Nr1d1-shRNA and Scr-shRNA groups.
Primer sequences for target and reference genes
Microarray gene expression analysis of NAc with knockdown of Nr1d1 in females and males
Pathway analysis in females with NIH DAVID, ToppCluster and MSET
L1000CDS identification of drugs that mimic up and down regulated genes in female Nr1d1 knockdown array
Acknowledgements
This work was supported by the National Institutes of Health Grant R21 MH 109714 to SC Gammie. The authors wish to thank Corinna Burger and Sue Osting for technical assistance with microinjection of AAV vectors, JaeJung Kim and the University of Chicago Genomics Core for microarray technical assistance, and Kate Skogan and Jeff Alexander for animal care.
Abbreviations
- AAV
adeno-associated virus
- Baml1
aryl hydrocarbon receptor nuclear translocator-like
- Bcl2
B cell leukemia/lymphoma 2
- BPD
bipolar disorder
- Cacna1h
calcium channel, voltage-dependent, T type, alpha 1H subunit
- Cirbp
cold inducible RNA binding protein
- Clock
circadian locomotor output cycles kaput
- CPP
conditioned place preference
- CPu
caudate putamen
- Cry1
cryptochrome 1 (photolyase-like)
- CTX
cerebral cortex
- DAVID
the database for annotation, visualization and integrated discovery
- Drd1a
dopamine receptor D1
- Eny2
ENY2 transcription and export complex 2 subunit
- Ethe1
ethylmalonic encephalopathy 1
- Fabp5
fatty acid-binding protein, epidermal
- Foxn2
forkhead box N2
- Frmd4a
FERM domain containing 4A
- Gabra2
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2
- GEO
gene expression omnibus
- GFP
green fluorescent protein
- Grin2b
glutamate receptor, ionotropic, NMDA2B (epsilon 2)
- H1f0
H1 histone family, member 0
- HDAC
histone deacetylase
- Hdac3
histone deacetylase 3
- Hdac4
histone deacetylase 4
- Hdac6
histone deacetylase 6
- Hif1a
hypoxia inducible factor 1, alpha subunit
- Hnrnpu
heterogeneous nuclear ribonucleoprotein U
- LINCS
the library of integrated network-based cellular signatures
- Morf4l1
mortality factor 4 like 1
- MSET
modular single-set enrichment test
- NAc
nucleus accumbens
- Ncoa3
nuclear receptor coactivator 3
- Nfil3
nuclear factor, interleukin 3, regulated
- Nr1d1
nuclear receptor subfamily 1, group D, member 1
- Oprd1
opioid receptor, delta 1
- PCA
principal component analysis
- Pdyn
prodynorphin
- Penk
preproenkephalin
- Per1
period circadian clock 1
- Per2
period circadian clock 2
- Pgr
progesterone receptor
- Phf7
PHD finger protein 7
- qPCR
quantitative real-time PCR
- RMA
robust multi-array average
- Scr-shRNA
scrambled short hairpin RNA
- shRNA
short hairpin RNA
- Suds3
suppressor of defective silencing 3 homolog
- TAC
transcriptome analysis console
- Trim28
tripartite motif-containing 28
- Tsc2
tuberous sclerosis 2
- WGCNA
weighted gene co-expression network analysis
- Yeats4
YEATS domain containing 4
- Ywhaz
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
- Zcrb1
zinc finger CCHC-type and RNA binding motif 1
- ZT
zeitgeber time
Footnotes
Competing interests
The authors declare that there was no commercial or financial relationship that could be considered a potential conflict of interest.
Data accessibility
Data will be available upon request from the corresponding author.
Reference
- Banerjee S, Wang Y, Solt LA, Griffett K, Kazantzis M, Amador A, El-Gendy BM, Huitron-Resendiz S, Roberts AJ, Shin Y, Kamenecka TM & Burris TP (2014) Pharmacological targeting of the mammalian clock regulates sleep architecture and emotional behaviour. Nat Commun, 5, 5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barresi V, Spampinato G, Musso N, Trovato Salinaro A, Rizzarelli E & Condorelli DF (2016) ATOX1 gene silencing increases susceptibility to anticancer therapy based on copper ionophores or chelating drugs. J. Inorg. Biochem, 156, 145–152. [DOI] [PubMed] [Google Scholar]
- Bevins RA & Bardo MT (1999) Conditioned increase in place preference by access to novel objects: antagonism by MK-801. Behav. Brain Res, 99, 53–60. [DOI] [PubMed] [Google Scholar]
- Burger C, Gorbatyuk OS, Velardo MJ, Peden CS, Williams P, Zolotukhin S, Reier PJ, Mandel RJ & Muzyczka N (2004) Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol. Ther, 10, 302–317. [DOI] [PubMed] [Google Scholar]
- Can A, Dao DT, Arad M, Terrillion CE, Piantadosi SC & Gould TD (2012) The mouse forced swim test. J Vis Exp, e3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlezon WA Jr. & Thomas MJ (2009) Biological substrates of reward and aversion: a nucleus accumbens activity hypothesis. Neuropharmacology, 56 Suppl 1, 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DL, Davis JF, Magrisso IJ, Fitzgerald ME, Lipton JW & Benoit SC (2012) Orexin signaling in the paraventricular thalamic nucleus modulates mesolimbic dopamine and hedonic feeding in the rat. Neuroscience, 210, 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Lee EJ, Yun S, Choe HK, Park SB, Son HJ, Kim KS, Dluzen DE, Lee I, Hwang O, Son GH & Kim K (2014) Impact of circadian nuclear receptor REV-ERBalpha on midbrain dopamine production and mood regulation. Cell, 157, 858–868. [DOI] [PubMed] [Google Scholar]
- Cordes MA, Stevenson SA, Driessen TM, Eisinger BE & Riters LV (2015) Sexually-motivated song is predicted by androgen-and opioid-related gene expression in the medial preoptic nucleus of male European starlings (Sturnus vulgaris). Behav. Brain Res, 278, 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington HE 3rd, Maze I, LaPlant QC, Vialou VF, Ohnishi YN, Berton O, Fass DM, Renthal W, Rush AJ 3rd, Wu EY, Ghose S, Krishnan V, Russo SJ, Tamminga C, Haggarty SJ & Nestler EJ (2009) Antidepressant actions of histone deacetylase inhibitors. J. Neurosci, 29, 11451–11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson BL, Stein CS, Heth JA, Martins I, Kotin RM, Derksen TA, Zabner J, Ghodsi A & Chiorini JA (2000) Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc. Natl. Acad. Sci. U. S. A, 97, 3428–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Solis CA, Hosek MP, Holehonnur R, Ho A, Banerjee A, Luong JA, Jones LE, Chaturvedi D & Ploski JE (2017) Adeno-associated viral serotypes differentially transduce inhibitory neurons within the rat amygdala. Brain Res, 1672, 148–162. [DOI] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, Lathrop GM, Abecasis GR & Cookson WO (2007) A genome-wide association study of global gene expression. Nat. Genet, 39, 1202–1207. [DOI] [PubMed] [Google Scholar]
- Dolen G, Darvishzadeh A, Huang KW & Malenka RC (2013) Social reward requires coordinated activity of nucleus accumbens oxytocin and serotonin. Nature, 501, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas LA, Varlinskaya EI & Spear LP (2003) Novel-object place conditioning in adolescent and adult male and female rats: effects of social isolation. Physiol. Behav, 80, 317–325. [DOI] [PubMed] [Google Scholar]
- Driessen TM, Eisinger BE, Zhao C, Stevenson SA, Saul MC & Gammie SC (2014) Genes showing altered expression in the medial preoptic area in the highly social maternal phenotype are related to autism and other disorders with social deficits. BMC Neurosci, 15, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger BE, Driessen TM, Zhao C & Gammie SC (2014) Medial prefrontal cortex: genes linked to bipolar disorder and schizophrenia have altered expression in the highly social maternal phenotype. Front. Behav. Neurosci, 8, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger BE, Saul MC, Driessen TM & Gammie SC (2013a) Development of a versatile enrichment analysis tool reveals associations between the maternal brain and mental health disorders, including autism. BMC Neurosci., 14, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger BE, Zhao C, Driessen TM, Saul MC & Gammie SC (2013b) Large scale expression changes of genes related to neuronal signaling and developmental processes found in lateral septum of postpartum outbred mice. PLoS One, 8, e63824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hachem N, Ba-Alawi W, Smith I, Mer AS & Haibe-Kains B (2017) Integrative cancer pharmacogenomics to establish drug mechanism of action: drug repurposing. Pharmacogenomics, 18, 1469–1472. [DOI] [PubMed] [Google Scholar]
- Etain B, Milhiet V, Bellivier F & Leboyer M (2011) Genetics of circadian rhythms and mood spectrum disorders. Eur. Neuropsychopharmacol, 21 Suppl 4, S676–682. [DOI] [PubMed] [Google Scholar]
- Everett LJ & Lazar MA (2014) Nuclear receptor Rev-erbalpha: up, down, and all around. Trends Endocrinol. Metab, 25, 586–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon E & McClung CA (2009) A role for the circadian genes in drug addiction. Neuropharmacology, 56 Suppl 1, 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis TC, Chandra R, Friend DM, Finkel E, Dayrit G, Miranda J, Brooks JM, Iniguez SD, O’Donnell P, Kravitz A & Lobo MK (2015) Nucleus accumbens medium spiny neuron subtypes mediate depression-related outcomes to social defeat stress. Biol. Psychiatry, 77, 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie SC, Driessen TM, Zhao C, Saul MC & Eisinger BE (2016) Genetic and neuroendocrine regulation of the postpartum brain. Front. Neuroendocrinol, 42, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeders NE, Lane JD & Smith JE (1984) Self-administration of methionine enkephalin into the nucleus accumbens. Pharmacol. Biochem. Behav, 20, 451–455. [DOI] [PubMed] [Google Scholar]
- Goldstein TR, Miklowitz DJ & Mullen KL (2006) Social skills knowledge and performance among adolescents with bipolar disorder. Bipolar Disord, 8, 350–361. [DOI] [PubMed] [Google Scholar]
- Gopal YN, Arora TS & Van Dyke MW (2007) Parthenolide specifically depletes histone deacetylase 1 protein and induces cell death through ataxia telangiectasia mutated. Chem. Biol, 14, 813–823. [DOI] [PubMed] [Google Scholar]
- Goto M, Mizuno M, Matsumoto A, Yang Z, Jimbo EF, Tabata H, Yamagata T & Nagata KI (2017) Role of a circadian-relevant gene NR1D1 in brain development: possible involvement in the pathophysiology of autism spectrum disorders. Sci. Rep, 7, 43945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griebel G, Perrault G & Sanger DJ (1997) CCK receptor antagonists in animal models of anxiety: comparison between exploration tests, conflict procedures and a model based on defensive behaviours. Behav. Pharmacol, 8, 549–560. [DOI] [PubMed] [Google Scholar]
- Hafner H, Nowotny B, Loffler W, an der Heiden W & Maurer K (1995) When and how does schizophrenia produce social deficits? Eur. Arch. Psychiatry Clin. Neurosci, 246, 17–28. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Reddy AB & Maywood ES (2003) A clockwork web: circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci, 4, 649–661. [DOI] [PubMed] [Google Scholar]
- Hauser H & Gandelman R (1985) Lever pressing for pups: evidence for hormonal influence upon maternal behavior of mice. Horm. Behav, 19, 454–468. [DOI] [PubMed] [Google Scholar]
- Hommel JD, Sears RM, Georgescu D, Simmons DL & DiLeone RJ (2003) Local gene knockdown in the brain using viral-mediated RNA interference. Nat. Med, 9, 1539–1544. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R & Lempicki RA (2009) Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics, Chapter 13, Unit 13 11. [DOI] [PubMed] [Google Scholar]
- Ikemoto S, Glazier BS, Murphy JM & McBride WJ (1997) Role of dopamine D1 and D2 receptors in the nucleus accumbens in mediating reward. J. Neurosci, 17, 8580–8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR (2003) Is social attachment an addictive disorder? Physiol. Behav, 79, 351–357. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U & Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Kaimal V, Bardes EE, Tabar SC, Jegga AG & Aronow BJ (2010) ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res, 38, W96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kripke DF, Nievergelt CM, Joo E, Shekhtman T & Kelsoe JR (2009) Circadian polymorphisms associated with affective disorders. J Circadian Rhythms, 7, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronfeld-Schor N & Einat H (2012) Circadian rhythms and depression: human psychopathology and animal models. Neuropharmacology, 62, 101–114. [DOI] [PubMed] [Google Scholar]
- Langfelder P & Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kang S & Kim W (2016) Drug Repositioning for Cancer Therapy Based on Large-Scale Drug-Induced Transcriptional Signatures. PLoS One, 11, e0150460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinsohn PM, Gotlib IH, Lewinsohn M, Seeley JR & Allen NB (1998) Gender differences in anxiety disorders and anxiety symptoms in adolescents. J. Abnorm. Psychol, 107, 109–117. [DOI] [PubMed] [Google Scholar]
- Lewinsohn PM, Mischel W, Chaplin W & Barton R (1980) Social competence and depression: the role of illusory self-perceptions. J. Abnorm. Psychol, 89, 203–212. [DOI] [PubMed] [Google Scholar]
- Li J, Zhou D, Qiu W, Shi Y, Yang JJ, Chen S, Wang Q & Pan H (2018) Application of Weighted Gene Co-expression Network Analysis for Data from Paired Design. Sci. Rep, 8, 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lee J, Salazar Hernandez MA, Mazitschek R & Ozcan U (2015) Treatment of obesity with celastrol. Cell, 161, 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonstein JS & De Vries GJ (2000) Sex differences in the parental behavior of rodents. Neurosci. Biobehav. Rev, 24, 669–686. [DOI] [PubMed] [Google Scholar]
- Makino S, Gold PW & Schulkin J (1994) Effects of corticosterone on CRH mRNA and content in the bed nucleus of the stria terminalis; comparison with the effects in the central nucleus of the amygdala and the paraventricular nucleus of the hypothalamus. Brain Res, 657, 141–149. [DOI] [PubMed] [Google Scholar]
- Markakis EA, Vives KP, Bober J, Leichtle S, Leranth C, Beecham J, Elsworth JD, Roth RH, Samulski RJ & Redmond DE Jr. (2010) Comparative transduction efficiency of AAV vector serotypes 1–6 in the substantia nigra and striatum of the primate brain. Mol. Ther, 18, 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MR, Ehlert EM, Eggers R, Pool CW, Hermening S, Huseinovic A, Timmermans E, Blits B & Verhaagen J (2010) Comparison of AAV serotypes for gene delivery to dorsal root ganglion neurons. Mol. Ther, 18, 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson BJ & Morrell JI (2005) Preference for cocaine- versus pup-associated cues differentially activates neurons expressing either Fos or cocaine- and amphetamine-regulated transcript in lactating, maternal rodents. Neuroscience, 135, 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson BJ, Williams S, Rosenblatt JS & Morrell JI (2001) Comparison of two positive reinforcing stimuli: pups and cocaine throughout the postpartum period. Behav. Neurosci, 115, 683–694. [DOI] [PubMed] [Google Scholar]
- Mattson BJ, Williams SE, Rosenblatt JS & Morrell JI (2003) Preferences for cocaine- or pup-associated chambers differentiates otherwise behaviorally identical postpartum maternal rats. Psychopharmacology (Berl.), 167, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA (2007) Circadian genes, rhythms and the biology of mood disorders. Pharmacol. Ther, 114, 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan EM, Halfon O, Magistretti PJ & Cardinaux JR (2016) The HDAC inhibitor SAHA improves depressive-like behavior of CRTC1-deficient mice: Possible relevance for treatment-resistant depression. Neuropharmacology, 107, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, Olson SL, Turek FW, Levine JE, Horton TH & Takahashi JS (2004) Circadian Clock mutation disrupts estrous cyclicity and maintenance of pregnancy. Curr. Biol, 14, 1367–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk JA, Green CB & Takahashi JS (2012) Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci, 35, 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollema NJ, Yuan Y, Jelcick AS, Sachs AJ, von Alpen D, Schorderet D, Escher P & Haider NB (2011) Nuclear receptor Rev-erb alpha (Nr1d1) functions in concert with Nr2e3 to regulate transcriptional networks in the retina. PLoS One, 6, e17494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy P, Sigman M, Ungerer J & Sherman T (1986) Defining the social deficits of autism: the contribution of non-verbal communication measures. J. Child Psychol. Psychiatry, 27, 657–669. [DOI] [PubMed] [Google Scholar]
- Newton SS, Thome J, Wallace TL, Shirayama Y, Schlesinger L, Sakai N, Chen J, Neve R, Nestler EJ & Duman RS (2002) Inhibition of cAMP response element-binding protein or dynorphin in the nucleus accumbens produces an antidepressant-like effect. J. Neurosci, 22, 10883–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numan M, Roach JK, del Cerro MC, Guillamon A, Segovia S, Sheehan TP & Numan MJ (1999) Expression of intracellular progesterone receptors in rat brain during different reproductive states, and involvement in maternal behavior. Brain Res, 830, 358–371. [DOI] [PubMed] [Google Scholar]
- Paxinos G & Franklin KBJ (2001) The Mouse Brain in Stereotaxic Coordinates. Academic Press, San Diego. [Google Scholar]
- Pendaries V, Malaisse J, Pellerin L, Le Lamer M, Nachat R, Kezic S, Schmitt AM, Paul C, Poumay Y, Serre G & Simon M (2014) Knockdown of filaggrin in a three-dimensional reconstructed human epidermis impairs keratinocyte differentiation. J. Invest. Dermatol, 134, 2938–2946. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res, 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW & Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res, 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccinelli M & Wilkinson G (2000) Gender differences in depression. Critical review. Br. J. Psychiatry, 177, 486–492. [DOI] [PubMed] [Google Scholar]
- Powell TR, Murphy T, Lee SH, Price J, Thuret S & Breen G (2017) Transcriptomic profiling of human hippocampal progenitor cells treated with antidepressants and its application in drug repositioning. J Psychopharmacol, 31, 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U & Schibler U (2002) The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell, 110, 251–260. [DOI] [PubMed] [Google Scholar]
- Rath MF, Rohde K, Fahrenkrug J & Moller M (2013) Circadian clock components in the rat neocortex: daily dynamics, localization and regulation. Brain Struct Funct, 218, 551–562. [DOI] [PubMed] [Google Scholar]
- Rath MF, Rovsing L & Moller M (2014) Circadian oscillators in the mouse brain: molecular clock components in the neocortex and cerebellar cortex. Cell Tissue Res, 357, 743–755. [DOI] [PubMed] [Google Scholar]
- Raynor K, Kong H, Chen Y, Yasuda K, Yu L, Bell GI & Reisine T (1994) Pharmacological characterization of the cloned kappa-, delta-, and mu-opioid receptors. Mol. Pharmacol, 45, 330–334. [PubMed] [Google Scholar]
- Rinn JL & Snyder M (2005) Sexual dimorphism in mammalian gene expression. Trends Genet, 21, 298–305. [DOI] [PubMed] [Google Scholar]
- Saitoh A, Kimura Y, Suzuki T, Kawai K, Nagase H & Kamei J (2004) Potential anxiolytic and antidepressant-like activities of SNC80, a selective delta-opioid agonist, in behavioral models in rodents. J. Pharmacol. Sci, 95, 374–380. [DOI] [PubMed] [Google Scholar]
- Saul MC, Majdak P, Perez S, Reilly M, Garland T Jr. & Rhodes JS (2017) High motivation for exercise is associated with altered chromatin regulators of monoamine receptor gene expression in the striatum of selectively bred mice. Genes, brain, and behavior, 16, 328–341. [DOI] [PubMed] [Google Scholar]
- Scotti MA, Lee G, Stevenson SA, Ostromecki AM, Wied TJ, Kula DJ, Gessay GM & Gammie SC (2011) Behavioral and pharmacological assessment of a potential new mouse model for mania. Physiol. Behav, 103, 376–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan T & Numan M (2002) Estrogen, progesterone, and pregnancy termination alter neural activity in brain regions that control maternal behavior in rats. Neuroendocrinology, 75, 12–23. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Krebs S, Bauersachs S, Blum H, Wolf E & Miyamoto A (2010) Actions and interactions of progesterone and estrogen on transcriptome profiles of the bovine endometrium. Physiol. Genomics, 42A, 290–300. [DOI] [PubMed] [Google Scholar]
- Shirayama Y & Chaki S (2006) Neurochemistry of the nucleus accumbens and its relevance to depression and antidepressant action in rodents. Curr. Neuropharmacol, 4, 277–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C & Crawley JN (2010) Behavioural phenotyping assays for mouse models of autism. Nat. Rev. Neurosci, 11, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons D & Self DW (2009) Role of mu- and delta-opioid receptors in the nucleus accumbens in cocaine-seeking behavior. Neuropsychopharmacology, 34, 1946–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, Yoo SH, Takahashi JS, Butler AA, Kamenecka TM & Burris TP (2012) Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature, 485, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S, Falcon E, Kumar J, Krishnan V, Mukherjee S, Birnbaum SG & McClung CA (2013) Circadian genes Period 1 and Period 2 in the nucleus accumbens regulate anxiety-related behavior. Eur. J. Neurosci, 37, 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffon C, Adams J, van der Fits L, Wen S, Townsend P, Ganesan A, Hodges E, Vermeer M & Packham G (2011) The histone deacetylase inhibitors vorinostat and romidepsin downmodulate IL-10 expression in cutaneous T-cell lymphoma cells. Br. J. Pharmacol, 162, 1590–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torra IP, Tsibulsky V, Delaunay F, Saladin R, Laudet V, Fruchart JC, Kosykh V & Staels B (2000) Circadian and glucocorticoid regulation of Rev-erbalpha expression in liver. Endocrinology, 141, 3799–3806. [DOI] [PubMed] [Google Scholar]
- Trabzuni D, Ramasamy A, Imran S, Walker R, Smith C, Weale ME, Hardy J, Ryten M & North American Brain Expression, C. (2013) Widespread sex differences in gene expression and splicing in the adult human brain. Nat Commun, 4, 2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trezza V, Damsteegt R, Achterberg EJ & Vanderschuren LJ (2011) Nucleus accumbens mu-opioid receptors mediate social reward. J. Neurosci, 31, 6362–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M & Hashimoto S (2005) System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet, 37, 187–192. [DOI] [PubMed] [Google Scholar]
- Uz T, Ahmed R, Akhisaroglu M, Kurtuncu M, Imbesi M, Dirim Arslan A & Manev H (2005) Effect of fluoxetine and cocaine on the expression of clock genes in the mouse hippocampus and striatum. Neuroscience, 134, 1309–1316. [DOI] [PubMed] [Google Scholar]
- Wang J, Gutala R, Hwang YY, Kim JM, Konu O, Ma JZ & Li MD (2008) Strain- and region-specific gene expression profiles in mouse brain in response to chronic nicotine treatment. Genes, brain, and behavior, 7, 78–87. [DOI] [PubMed] [Google Scholar]
- Warner-Schmidt JL, Schmidt EF, Marshall JJ, Rubin AJ, Arango-Lievano M, Kaplitt MG, Ibanez-Tallon I, Heintz N & Greengard P (2012) Cholinergic interneurons in the nucleus accumbens regulate depression-like behavior. Proc. Natl. Acad. Sci. U. S. A, 109, 11360–11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Silverman JL & Crawley JN (2011) Automated three-chambered social approach task for mice. Curr. Protoc. Neurosci, Chapter 8, Unit 8 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrindast MR, Babapoor-Farrokhran S, Babapoor-Farrokhran S & Rezayof A (2008) Involvement of opioidergic system of the ventral hippocampus, the nucleus accumbens or the central amygdala in anxiety-related behavior. Life Sci, 82, 1175–1181. [DOI] [PubMed] [Google Scholar]
- Zhang B & Horvath S (2005) Ridge regression based hybrid genetic algorithms for multi-locus quantitative trait mapping. Int J Bioinform Res Appl, 1, 261–272. [DOI] [PubMed] [Google Scholar]
- Zhao C, Driessen T & Gammie SC (2012a) Glutamic acid decarboxylase 65 and 67 expression in the lateral septum is up-regulated in association with the postpartum period in mice. Brain Res, 1470, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Eisinger BE, Driessen TM & Gammie SC (2014) Addiction and reward-related genes show altered expression in the postpartum nucleus accumbens. Front. Behav. Neurosci, 8, 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Saul MC, Driessen T & Gammie SC (2012b) Gene expression changes in the septum: possible implications for microRNAs in sculpting the maternal brain. PLoS One, 7, e38602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C & Mallampalli RK (2014) Regulation of histone modifying enzymes by the ubiquitin-proteasome system. Biochim. Biophys. Acta, 1843, 694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Double immunofluorescence labeling of Nr1d1 (A) with NeuN (neuronal marker, B) showing that the vast majority of Nr1d1-expressing cells are neurons in the NAc of mouse brain. Double fluorescence labeling was carried out with a tyramide signal amplification (TSA) method. Sections were incubated overnight at 4°C with mouse anti-Nr1d1 primary antibodies (LifeSpan BioSciences, LS-B4487, diluted 1:2000). After primary antibody incubation, sections were washed and incubated for 1 hr with HRP-conjugated horse anti-mouse antiserum (Cell Signaling Technology; #7076, diluted 1:100). After several washes, they were incubated for 10 min in Cy3-conjugated tyramide (TSATM Plus Cyanine 3 kit, PerkinElmer, Waltham, MA; red for Nr1d1 labeling) by diluting TSA stock solution 1:50 in 1× Amplification Diluent. After washing with wash buffer, sections were incubated in heated citric acid buffer (10 mM, pH 6.0) at 95°C for 5 min to eliminate HRP activity from the initial TSA reaction and cross-reaction between IgGs raised in the mouse. Sections were then incubated overnight at 4°C with mouse anti-NeuN (Millipore, MAB377, diluted 1:000) primary antibodies. Sections were washed and incubated for 1 hr with HRP-conjugated horse anti-mouse antiserum. After 3 washes, they were incubated for 30 min in Alexa Fluor 488-conjugated tyramide (Molecular Probes, Eugene, OR; green for NeuN labeling) by diluting TSA stock solution 1:100 in 1×Amplification reagent. Following washing 3×10 min with wash buffer, sections were mounted onto slides using DePeX mounting medium (Serva, Heidelberg, Germany), air-dried and stored in the dark at 4°C. Note that the vast majority of Nr1d1-expressing cells were colocalized with NeuN (C), suggesting that they are neurons. Arrows indicate the typical examples of double-labeled neurons in the NAc. Scale bar = 50 μm.
Verification of the injection site and extent of viral transduction in mice injected with AAV vectors expressing Nr1d1 shRNA into the NAc. Immunofluorescence labeling with GFP antibody that serves as a reporter to label the transduced neurons was performed. Brain tissue was collected 3 weeks after stereotaxic injection of AAV Nr1d1 shRNA. Sections were incubated overnight at 4°C with rabbit anti-GFP primary antibodies (Molecular Probes, A-11122, diluted 1:1000). After primary antibody incubation, sections were washed and incubated for 2 hrs with Alexa Fluor 488-conjugated donkey anti-rabbit antiserum (Molecular Probes; A-21206, diluted 1:100). Two representative lower (A, B) and one higher (C) magnification photomicrographs show that the GFP-positive cells were predominantly expressed in the NAc, while sporadically distributed in the adjacent areas. Scale bars, 50 μm in A, B; 20 μm in C.
Quantitative real-time PCR analysis of Nr1d1 and GFP gene expression in the CPu and CTX of female and male mice. Relative expression distribution of mRNA (Y-axis) represented as a ratio of Nr1d1 shRNA-injected versus Scr shRNA-injected mice, was normalized against two reference genes Ywhaz and ?-actin, and shown by box-and-whisker plot as medians (dashed lines), interquartile range (boxes) and ranges (whiskers). Note that no significant differences were observed in mRNAs of Nr1d1 and GFP in CPu and CTX between the Nr1d1-shRNA and Scr-shRNA groups.
Primer sequences for target and reference genes
Microarray gene expression analysis of NAc with knockdown of Nr1d1 in females and males
Pathway analysis in females with NIH DAVID, ToppCluster and MSET
L1000CDS identification of drugs that mimic up and down regulated genes in female Nr1d1 knockdown array