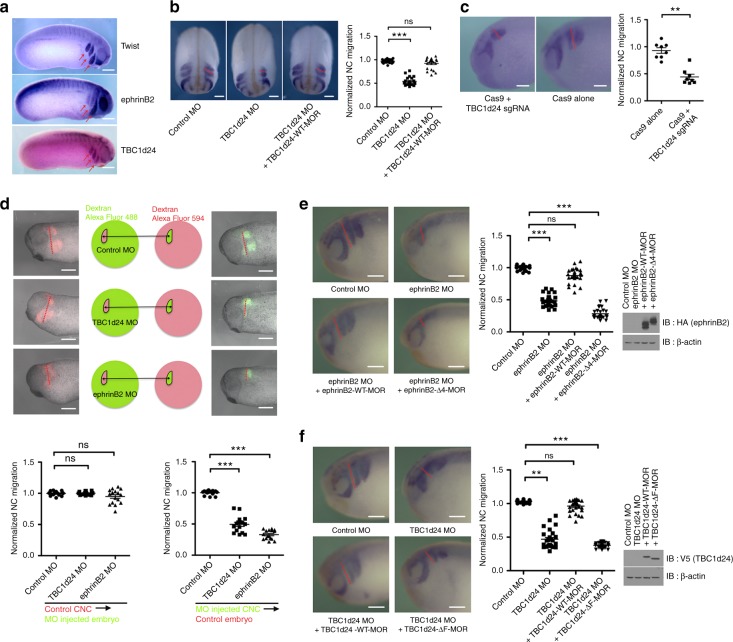
Fig. 4.
EphrinB2–TBC1d24 interaction regulates CNC cell migration. a Spatial expression pattern of Twist, ephrinB2 and TBC1d24. WISH of stage 24 embryos with probes for twist (a CNC marker), ephrinB2 and TBC1d24 (red arrows indicate CNC). Lateral view, anterior is right. b WISH using the Twist antisense probe in embryos injected with Control MO or TBC1d24MO alone or along with TBC1d24 MO-resistant RNA. MOs and/or RNAs were injected into the D.1.2 blastomere. The dotted red line denotes distance of CNC migration on the injected side of the tailbud stage embryo. Anterior/dorsal view. Quantification with non-parametric ANOVA (Kruskal–Wallis test), P < 0.0001. c WISH using the Twist antisense probe in F0 embryos injected with sgRNA for TBC1d24 and/or Cas9 protein. Dotted red line denotes distance of CNC migration. Quantification with Mann–Whitney test: **P = 0.0013, two-tailed). d Green fluorescein dextran (GFD)-labelled neural crest explant tissue from control, TBC1d24 or ephrinB2-depleted embryos was exchanged with normal neural crest explant tissue from red fluorescein dextran (RFD)-labelled embryos at stage 20 as indicated in the schematic (left). At the tailbud stage (right), it is observed that depletion of ephrinB2 or TBC1d24 causes neural crest cell migration defects in a tissue-autonomous manner. Quantification with one-way ANOVA, P < 0.0001. e WISH using the Twist antisense probe in the embryos injected with ephrinB2 MO alone or with MO-resistant wild-type or Δ4 ephrinB2 mutant RNA as indicated. Dotted red line denotes distance of CNC migration. Quantification with non-parametric ANOVA (Kruskal–Wallis test), P < 0.0001. Western blot shows the similar indicated protein expression. f WISH using twist probe in embryos injected with the indicated MOs and RNAs. The red arrow indicates the CNC migration differences between embryos. Western analysis shows that mutant and wild-type MO-resistant RNAs yield similar levels of protein expression. Quantification with non-parametric ANOVA (Kruskal–Wallis test), P < 0.0001; Bar, 200 μm. All histograms and scatterplots represent mean ± s.e.m from three biological repeats; Dunnett’s multiple comparison; **P < 0.01 and ***P < 0.001, ns: no statistical differences between groups