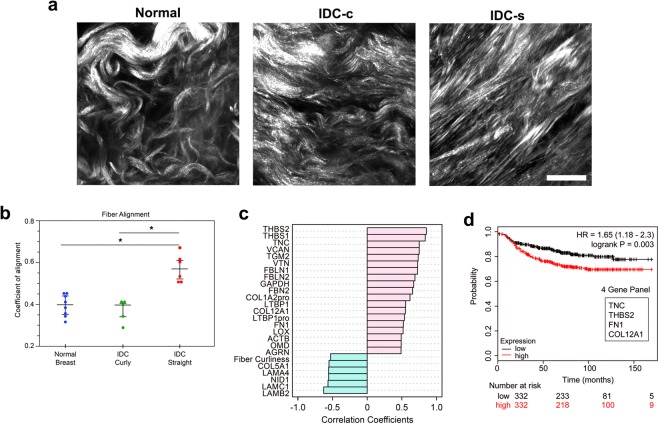
Figure 2.
Collagen fiber orientation analysis identifies ECM proteins that correlate with collagen fiber alignment, not simply progression to IDC. (a) Representative second-harmonic generation (SHG) images of normal breast and IDC tissues. The normal breast tissues always exhibited curly/wavy, randomly organized collagen (left), whereas the IDC samples showed more variability. Some of the IDC samples contained the randomly organized collagen similar to normal breast (middle), while others possessed straightened/aligned collagen architectures, similar to TACS-3 (right). Images presented here are compressed z-series, ‘max intensity.’ Scale bar = 25 µm. (b) Box plots of ct-FIRE and CurveAlign analysis for all SHG images of patient samples used in this study. Coefficient of alignment ranges from completely random fiber orientation, 0.0, to completely aligned fiber orientation, 1.0. (c) Pattern Analysis of log2-normalized, quantitative and global proteomics results to fiber alignment (normal breast, n = 8; IDC, n = 11). (d) Four increased ECM proteins for IDC/normal breast (collagen XII, COL12A1; fibronectin, FN; tenascin-C, TNC; and thrombospondin-2, THBS-2) formed a heightened correlative signature for poor distant metastasis-free survival in silico, using the K-M plotter database, than any ECM protein individually had correlated.