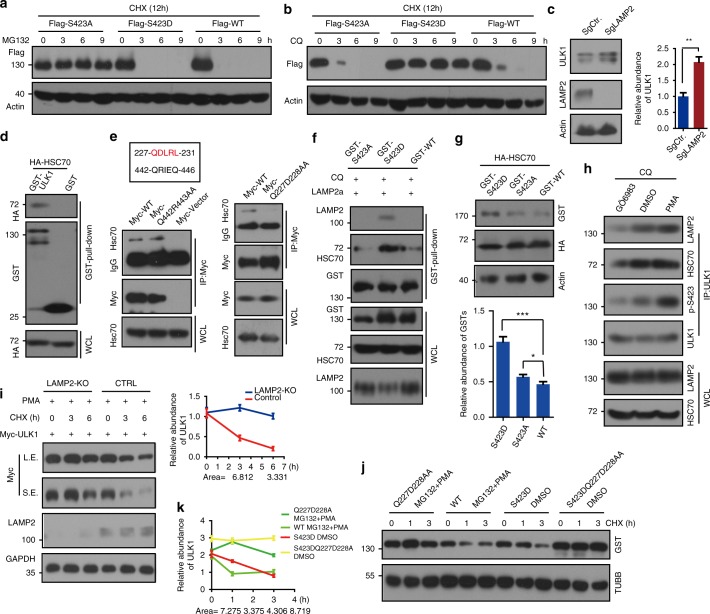
Fig. 6.
Phosphorylation of ULK1 is degraded through chaperone-mediated autophagy. a MG132 (proteasome inhibitor, 20 μm) prevents ULK1 S423A degradation but not degradation of ULK1 WT or S423D. Stable ULK1 WT and mutant HeLa cells were pretreated with cycloheximide (CHX, 20 μg mL−1), an inhibitor of protein synthesis, for 3 h. Then, the cells were treated with MG132 for a time course in the presence of CHX. The remaining ULK1 was detected by WB. b CQ inhibits ULK1 S423D degradation. The procedure is similar to Fig. 6a, but MG132 is replaced by CQ. c Accumulation of more ULK1 in Lamp2-KO cells. The abundance of ULK1 was quantified. d The interaction of ULK1 and HSC70 was determined by pull-down assay with overexpression of GST-ULK1 and HA-HSC70. e The HSC70 binding motif of ULK1 is between Q227 to L231, and the result was verified by double mutations and semi-endogenous co-IP assay by HSC70 antibody. f ULK1 S423D binds more HSC70 and LAMP2a than its counter partners. g Overexpression of HSC70 promotes ULK1 WT and S423Ds degradation, not S423A degradation. h PKCα activity increased the affinity of ULK1 to HSC70 and LAMP2. The results were detected with the indicated antibodies. WCL whole-cell lysate. i ULK1 degraded slowly in LAMP2-KO cells with PMA (a PKC activator) treatment. Myc-tagged ULK1 was transfected into the above two cell lines with the indicated drugs and time course. The remaining ULK1 was quantified. j Mutant ULK1 Q227D228AA prevents p-ULK1 degradation through the lysosome. k The quantification of Fig. 6j. All values are means ± SEM of three independent experiments: *p < 0.05, **p < 0.01, ***p < 0.001. WCL: whole-cell lysate, L.E.: long exposure, S.E.: short exposure