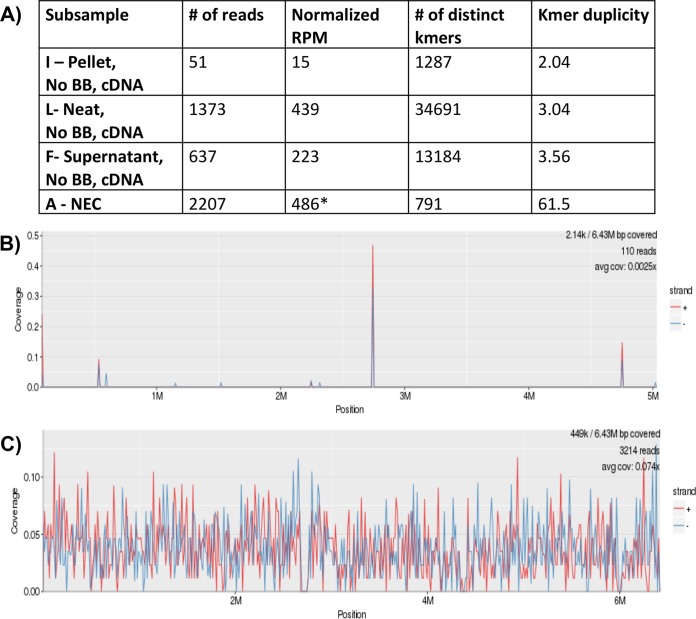
FIG 2.
Unique kmer and alignment analysis methods for CSF sample 3 positive for Pseudomonas aeruginosa. (A) Comparison of the numbers of unique kmers and kmer duplicity assigned to the P. aeruginosa reads among sample 3 cDNA subsamples (F, I, and L) with those of the negative extraction control (subsample A-NEC) by KrakenHLL. (B and C) Demonstration of the reads aligned to the P. aeruginosa genome found in the NEC (B) compared to those in the neat CSF sample without bead beating (BB) using cDNA methods (subsample L with 7.4% genome coverage) (C). An asterisk indicates a positive cutoff for these samples of 4,855 (10 times the RPM of the NEC).