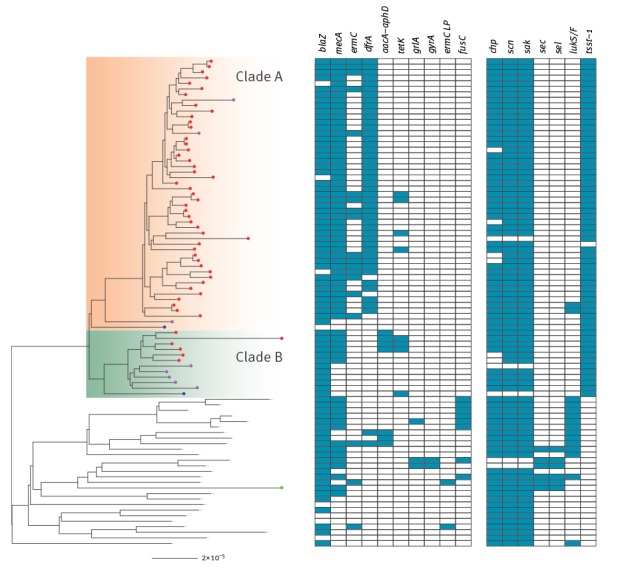
Figure 2.
Presence and absence of genes involved in antibiotic resistance and virulence for meticillin-resistant Staphylococcus aureus ST22 non-ST22-A populations, Gaza, 2009 (n = 88)
MRSA: meticillin-resistant Staphylococcus aureus; MSSA: meticillin-sensitive Staphylococcus aureus.
Left panel: midpoint-rooted maximum likelihood phylogenetic tree of Gaza isolates along with non-ST22-A isolates. Red tips: MRSA isolates from the Gaza region; purple tips: MSSA isolates from the Gaza region; green tip: strain from Bat Yam, Israel; blue tips: the two closest non-ST22-A isolates from Germany and Sudan.
Right panel: presence (blue) or absence (white) of genes.
Resistance genes: blaZ (penicillin), mecA (oxacillin), ermC (erythromycin), dfrA (trimethoprim), aacA-aphD (gentamicin), tetK (tetracycline), grlA and gyrA (fluoroquinolone), ermC LP (clindamycin), fusC (fusidic acid).
Toxin genes: chp (CHIPS), scn (SCIN), sak (staphylokinase), sec (enterotoxin C), sel (enterotoxin L), lukS/F-PV (Panton–Valentine leukocidin), tsst-1 (toxic shock syndrome toxin-1).