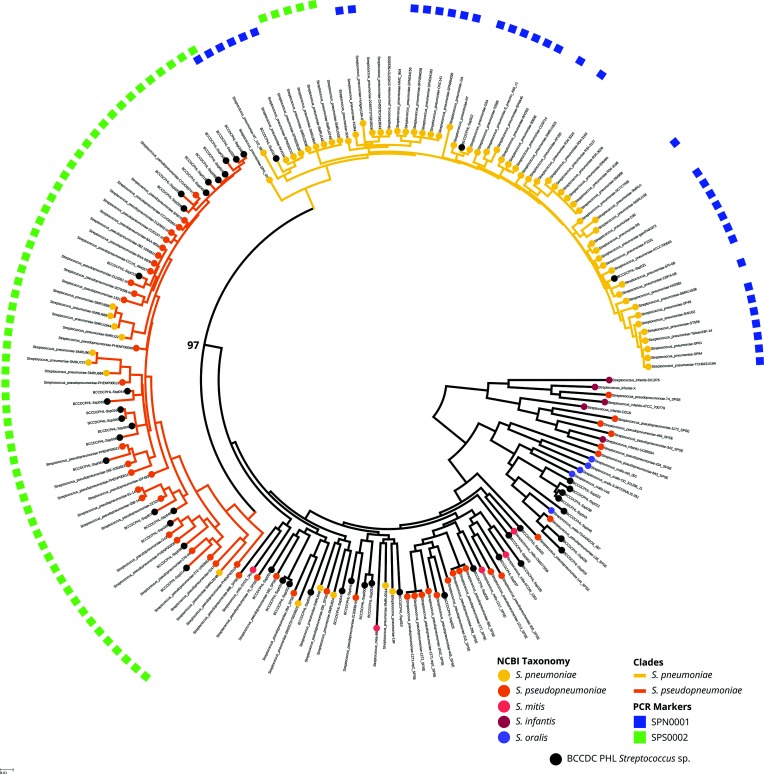
Fig. 2.
Phylogenetic tree of members of the genus Streptococcus and the presence/absence of the SPN0001 and SPS0002 PCR marker. A phylogenetic tree was reconstructed to show the relationship of the SPN0001 S. pneumoniae (blue squares) and SPS0002 S. pseudopneumoniae (green squares) PCR markers and the placement of the genomes on the tree. Note the S. gordonii, and S. australis isolates were excluded from this tree. S. pneumoniae (gold nodes); S. pseudopneumoniae (orange nodes); S. mitis (coral nodes); S. oralis (mauve nodes); S. infantis (magenta nodes); and BC Streptococcus sp. (black nodes). Bootstrap support value is indicated in black bold type at the node that separates the S. pneumoniae and S. pseudopneumoniae clades. All SPN0001-positive genomes clustered in the S. pneumoniae clade (yellow branches) while almost all SPS0002-positive genomes clustered in the S. pseudopneumoniae clade (orange branches). The three BC S. pneumoniae clinical isolates clustered with the other S. pneumoniae, while 22 BC SPS0002-positive Streptoccocus sp. clustered with the S. pseudopneumoniae clade. This tree also shows all discrepant results (excluding non-Streptococcus organisms and S. salivarius), such as SPN0001-negative S. pneumoniae and SPS0002-positive S. pneumoniae. Note that only RefSeq complete S. pneumoniae and discrepant S. pneumoniae genomes are shown as a representation of all 8051 S. pneumoniae genomes in RefSeq. Overall, 8027 out of 8051 S. pneumoniae were SPN0001-positive.