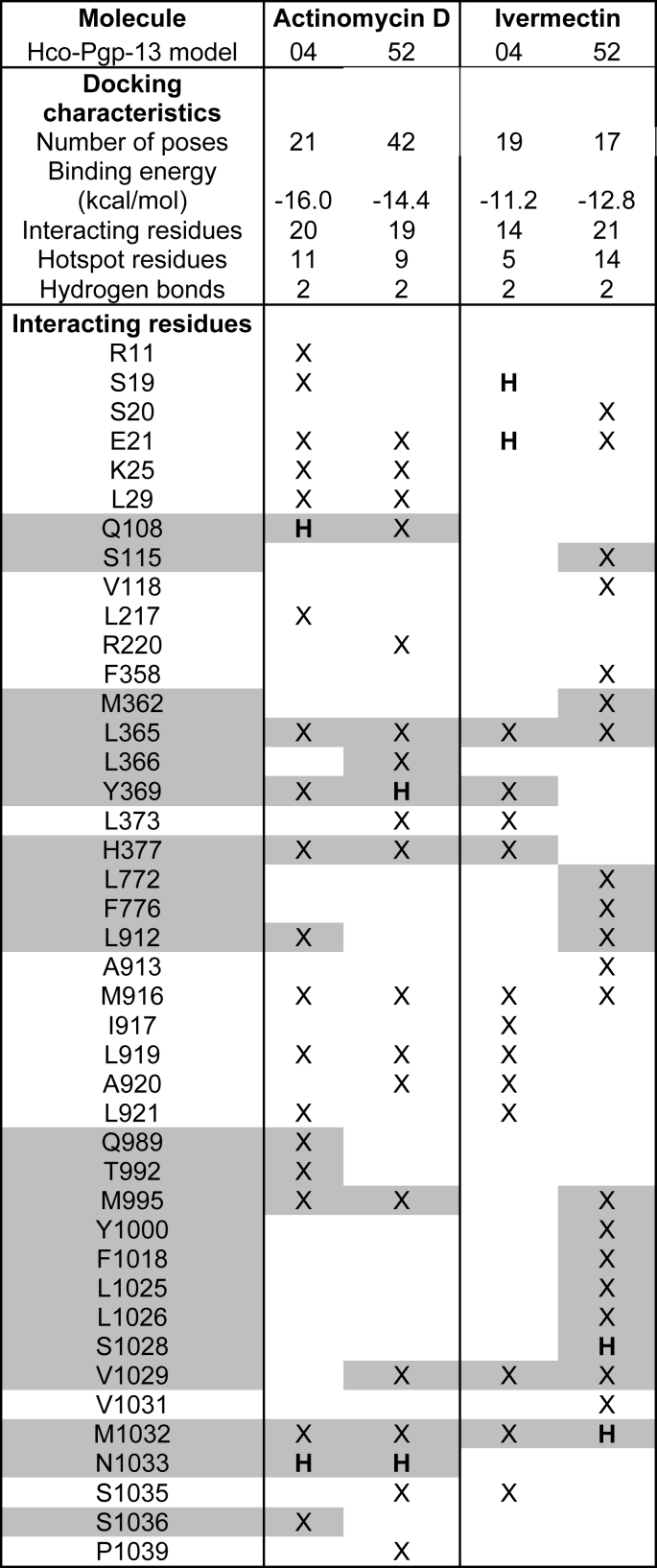
Table 2.
Docking characteristics and list of interacting residues of actinomycin D and ivermectin in the Hco-Pgp-13 structure. The parameters and interacting residues listed in column 1 stand for the lowest energy cluster of ACD on Hco-Pgp-13_04 (column 2) and Hco-Pgp-13_52 (column 3), and of IVM on Hco-Pgp-13_04 (column 4) and Hco-Pgp-13_52 (column 5). X: contact residues;  : hotspot residues. H: residues establishing a hydrogen bond.
: hotspot residues. H: residues establishing a hydrogen bond.
Legend for docking features: a) Number of poses: number of poses in the lowest energy cluster of histogram of poses clustered at rmsd = 2 Å. b) Binding energy (kcal/mol): the energy score calculated by Autodock 4.2. c) Interacting residues: number of contact residues as determined by AutoDock Tools graphical interface. d) Hotspot residues: number of contact residues identified as hotspot residues according to our determination listed in Table S3. e) Hydrogen bonds: number of H-bonds identified by AutoDock Tools between the docked ligands and the protein receptor.