Abstract
With the discovery that the level of RNA synthesis in human cells far exceeds what is required to express protein-coding genes, there has been a concerted scientific effort to identify, catalogue and uncover the biological functions of the non-coding transcriptome. Long, non-coding RNAs (lncRNAs) are a diverse group of RNAs with equally wide-ranging biological roles in the cell. An increasing number of studies have reported alterations in the expression of lncRNAs in various cancers, although unravelling how they contribute specifically to the disease is a bigger challenge. Originally described as a brain-specific, non-coding RNA, BC200 (BCYRN1) is a 200-nucleotide, predominantly cytoplasmic lncRNA that has been linked to neurodegenerative disease and several types of cancer. Here we summarise what is known about BC200, primarily from studies in neuronal systems, before turning to a review of recent work that aims to understand how this lncRNA contributes to cancer initiation, progression and metastasis, along with its possible clinical utility as a biomarker or therapeutic target.
Keywords: Long non-coding RNA, lncRNA, Cancer, BC200, BCYRN1, Translational regulation, RNA-protein interactions
1. Introduction
Over the past ten years, advances in sequencing technologies have led to the discovery that the human genome is widely transcribed into RNA; however, only 2% of the RNA produced is translated into proteins [1,2]. This means that the cellular contribution of the non-coding transcriptome is expanding. In particular, long, non-coding RNAs (lncRNAs) have emerged as regulators of normal cellular events and various disease states, including cancer [3,4].
As one class of non-coding RNAs, lncRNAs are defined as transcripts greater than 200 nucleotides and containing little or no protein-coding potential. The size criterion distinguishes lncRNAs from smaller, non-coding RNAs, such as microRNAs (miRNAs) and piwi-interacting RNAs (piRNAs) [5,6]. Studies indicate that most lncRNAs are synthesised by RNA polymerase II (Pol II) and are processed similarly to messenger RNAs (mRNAs), with capping, splicing and polyadenylation [7]. However, some lncRNAs, like Pol II transcript metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), have unique processing events that distinguish them from mRNAs [8,9]. In addition to Pol II transcription, some lncRNAs are transcribed by RNA polymerase III (Pol III) [10,11], adding another level of transcriptional complexity to this class of non-coding RNAs.
Not surprisingly, much scientific effort has been directed towards the discovery and annotation of lncRNAs in humans [12,13]. This has subsequently led to a proliferation of specialized lncRNA databases, such as NONCODEv5 [14] that integrates data from the genomes of 17 different species and LNCipedia [15] that contains close to 150,000 human annotated lncRNAs.
With tens of thousands of lncRNAs in human cells [14,16], there are significant challenges to understand their cellular roles, particularly as lncRNAs are often expressed at lower levels than mRNAs and exhibit poor sequence conservation across various species [17]. However, lack of sequence conservation does not equate to lack of function [18]. In recent years, significant progress has been made in characterising biological functions of lncRNAs. There are now many examples of lncRNAs affecting cellular processes like development, differentiation and proliferation, in addition to their impact at every level of gene expression from chromatin remodeling to translation [3,4,9,[19], [20], [21], [22]].
This review will focus on BC200, an lncRNA that was first discovered in primate brain in 1987 [23]. Conserved in humans [24], initial studies on human BC200 were focused on its neural-specific, dendritic localisation [25]. A link to cancer was provided a few years later by a key study revealing BC200 RNA expression in a number of human tumours by Northern blot analysis and in situ hybridization [26]. In that study, the authors found BC200 expression to be substantially increased in certain tumours, like breast, cervix, oesophagus, lung, ovary, parotid and tongue, while it is undetectable in corresponding normal tissues and in other cancers, such as bladder, colon and liver [26]. More recent investigations propose that BC200 has roles in cell migration, proliferation and survival [[27], [28], [29], [30], [31], [32], [33], [34]] – all suggesting that BC200 contributes to cancer development and progression.
In this review, we outline the general features of BC200 lncRNA, including its transcription, sequence elements and expression patterns. We then refer to BC200's evolutionary emergence and detail its analogues. To better understand BC200's functional significance, we present current knowledge of the RNA's protein interactions within the cell, highlighting several recent publications. Lastly, BC200's association with human disease, focusing on cancer, is discussed, along with the use of BC200 as a cancer diagnostic or prognostic biomarker.
2. Overview of BC200 lncRNA
2.1. Features of BC200 from transcription to cellular localisation
2.1.1. BC200 biosynthesis
BC200, also known as brain cytoplasmic RNA 1 (BCYRN1), is a non-coding RNA, sense-transcribed from its gene locus situated on human chromosome 2 between the protein-coding genes for calmodulin 2 (CALM2) and epithelial cellular adhesion molecule (EPCAM) [35,36]. BC200 was the first example of a primate, tissue-specific Pol III transcript [37], altering the view that Pol III was only responsible for the synthesis of transfer RNA (tRNA) and 5 S ribosomal RNA (rRNA) [10,11].
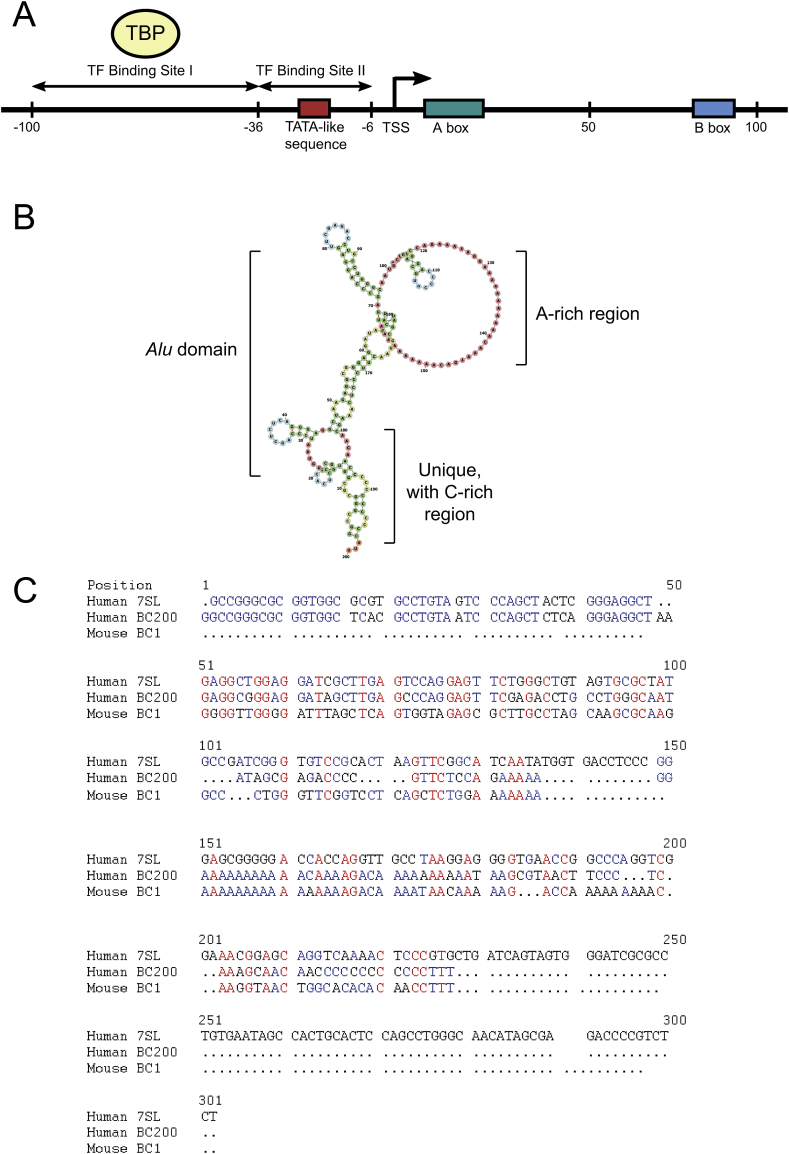
Recent work from Kim et al. (2017) offers an in-depth analysis of BC200 biosynthesis [38]. Prior to this study, analysis of the BC200 promoter region included the identification of a putative TATA box and two internal A and B box promoter elements [39], which are common features of Pol III promoters [11,40]. With their study, Kim et al. confirmed the critical role of the TATA box and internal promoter elements for BC200 transcription. In addition, the authors demonstrated that the 100-base pair (bp) region upstream to BC200 transcription start site (TSS) is characterized by two distinct regions: −100 to −36 bp, which is bound by transcription factors independent to the internal promoter elements, and a −35 to −6 bp region in which the binding of transcription factors is dependent on the A and B box elements (Fig. 1A).
Fig. 1.
Schematic representation of human BC200 promoter region, secondary structure and sequence similarity to 7SL and BC 1 RNAs.
(A) The BC200 promoter is divided into three domains. The first is transcription factor (TF) binding site I located between position −100 and −36 bp. The second is TF binding site II located between −35 bp and −6 bp, which contains a TATA box. The binding of TFs to this region is dependent on the third domain, containing two internal promoter elements, A box and B box. TATA binding protein (TBP) binds to the BC200 promoter in the region located between −100 bp to 30 bp [38]. Additional transcription factors that influence the BC200 promoter are not shown. (B) Folding parameters of BC200 RNA (accession number NR038088.1) were determined by mFold [156] and used in forna, an RNA secondary structure visualization tool [157]. Structural elements are colour-coded as follows: Green – stems (canonical helices); red – multiloops/junctions; yellow – interior loops; blue – hairpin loops; orange – 3′ unpaired region. The Alu-like, A-rich and unique regions of BC200 are indicated. (C) Pairwise alignment of BC200 with 7SL (human; accession number X04248.1) and BC1 (mouse; accession number NR001568.1) using MultiAlin [158]. Nucleotides are coloured blue if they are identical between two of the three RNAs; red indicates identical nucleotides among the three non-coding RNAs.
The authors further revealed that TATA binding protein (TBP) binds to the −100 bp region in human cervical carcinoma (HeLa) cells and that a deletion of the TATA box leads to an impaired BC200 transcription [38]. Indeed the TATA box and TBP-binding seems to be part of a conserved core promoter that is required for all eukaryotic polymerases [41].
While core promoter elements are essential for basal transcription, further levels of transcriptional regulation can be achieved by specific transcription factors binding to promoter elements and influencing transcriptional output. With regard to BC200 synthesis, two studies have examined the influence of specific transcription factors, c-MYC [27] and estrogen receptor α (ERα) [28], in lung and breast cancer cell lines respectively. Given that the MYC proto-oncogene is deregulated in many cancers [42,43], it remains to be seen if MYC drives BC200 expression in other cancers besides non-small cell lung cancer (NSCLC) [27]. Like other promoters, the specific transcription factors that regulate BC200 transcription at any given time will likely be dependent on cell type and context. Therefore, in addition to experimentally identifying the members of BC200 transcriptional network, it will be important to understand how the network is integrated and regulated within a wider cellular environment. Certainly there is still much to be discovered about the specific details BC200 biosynthesis, particularly in diseased cells.
2.1.2. BC200: structural elements and cellular localisation
Named after the length of its unique single exon of 200 nucleotides [23,25], sequence analysis of BC200 RNA sequence revealed three distinct sequence domains: a 5′ Alu element, a central adenosine-rich region and a 3′, 43-nucleotide, unique region containing a cytosine-rich stretch [25] (Fig. 1B).
Unlike many lncRNAs that remain and function in the nucleus, BC200 is classified as a small, cytoplasmic RNA (Ensembl release 90) [44]. Importantly, many of its molecular interactions involve proteins located in the cytoplasm (described in the following section). Initial evidence for its cytoplasmic location was based on its presence in the cytoplasmic, poly (A)+ RNA fraction of monkey and human brain samples [23]. More recent experiments have involved cell fractionation followed by quantitative, reverse transcription, polymerase chain reaction (qRT-PCR) to monitor relative expression levels of BC200 in nuclear versus cytoplasmic fractions [28,30]. In these experiments, BC200 is primarily present in the cytoplasm; however, it should be pointed out that there is a small, but detectable amount of BC200 in the nuclear fraction as well [28,30]. In an alternative approach using an antibody directed against BC200 RNA, Shin et al. (2017) observed punctate staining in both the cytoplasm and nucleus of cervical carcinoma cells and co-localisation of BC200 with proteins in processing bodies (P bodies) [45]. Taken together, BC200 is largely cytoplasmic; yet, with evidence that the RNA modulates alternative splicing of an apoptotic regulator protein, Bcl-x (B cell lymphoma 2 family member) [28], additional nuclear functions for BC200 cannot be ruled out.
2.1.3. BC200: confirmation of non-coding status
One surprising experimental observation in the lncRNA field has been that cytoplasmic lncRNAs are often found associated with translational machinery through ribosome profiling data [46,47], calling into the question their non-coding status. A study by Carlevaro-Fita et al. (2016) raises the possibility that the ribosome may play a role in regulating the degradation of cytoplasmic lncRNAs [48]. Interestingly, an increasing number of reports demonstrate that non-coding transcripts might in fact code for small peptides [46,49]. The identification of these peptide-coding, ‘non-coding’ RNAs remains difficult. As ribosome occupancy alone is not proof of the coding status [50], it still hints at the coding potential of the transcript. Indeed the line between lncRNAs and mRNAs is blurred, and it is not as clear cut as once was assumed [51,52]. In examining the coding potential of BC200 using published ribosome profiling data [[53], [54], [55], [56], [57]] in the GWIPS-viz Genome Browser from RiboGalaxy [58], we confirmed that BC200 does not associate with ribosomes, reflecting its true non-coding status as previously described [37].
2.1.4. BC200: tissue and cell-type expression
Indicative of its name, BC200 is found in the primate brain [23,25]. Among neural cells, BC200 is highly expressed in dendrites, where is it thought to play a role in local translational control [24,25,[59], [60], [61], [62]]. Given its expression in the nervous system, there have been numerous studies examining BC200's molecular interactions in neurons [[63], [64], [65], [66], [67], [68]], along with reports examining the precise function of BC200 [60,62]. The reader is referred to a recent review by Sosińska et al. [69] that comprehensively addresses what is known about BC200 in the regulation of neuronal gene expression.
Outside of neural tissue, BC200 is expressed in testes and ovary [30]; however, for this review, our focus is on BC200's link to various cancers. While BC200 is present in non-neuronal immortalized cell lines [30], it was a seminal study by Chen at al. that provided the first evidence of BC200 expression in various tumour types [70]. This was followed by the work of Iacoangeli et al. who examined BC200 specifically in invasive and pre-invasive breast cancer [71].
Since there is existing information about BC200, its interacting proteins and proposed mechanisms of action from neuronal systems, it is possible that there are mechanistic overlaps in neoplastic cells. However, recent work by Singh et al. [28] provides the first evidence of BC200's role in alternative splicing in breast cancer cells, something not observed previously in neurons. Therefore, BC200 might have roles in cancer initiation and/or progression that cannot be anticipated from previous studies.
2.2. History of BC200
2.2.1. Evolution of BC200
Unlike protein-coding RNAs, lncRNAs have limited sequence conservation among different species. This has caused significant debate because the lack of sequence conservation has led to the idea that lncRNAs are simply “transcriptional noise” [72]. Combined with challenges to test the functionality of many lncRNAs in homologous animal model systems [73], the biological functionality of the majority of lncRNAs remains to be discovered and robustly validated.
To better understand how lncRNAs became part of the human transcriptome, several mechanisms for the evolution of lncRNAs have been proposed [74]. Of those, one way that lncRNAs can emerge is by insertion of a transposable element into the genome that then becomes a functional transcriptional locus [74,75]. The first report of monkey BC200 highlights its sequence identity with a human Alu monomer [76] and refers to a retropositional event that led to BC200 [23].
By comparing the human BC200 (BCYRN1) gene sequence with the orthologous loci of the prosimians – galago, lemur and tarsier, Kuryshev et al. established that BC200 is absent from prosimian lineages [77]. They proposed that the BC200 gene arose between 35 and 55 million years ago, due to the insertion of a monomeric Alu element in the lineage leading to primates (Anthropoidea) [77]. Through successive events of substitutions and insertions, the gene continued to evolve until the Homininae subfamily, composed of gorillas, chimpanzees and humans [77].
2.2.2. BC200 and 7SL RNA: a common Alu element
From its initial characterisation by Tiedge et al., in 1993, it was shown that human BC200 shared sequence similarity to 7SL RNA, the 300-nucleotide, non-coding RNA that forms the signal recognition particle (SRP) [25,78]. As mentioned earlier, BC200 has a 5′ Alu element that has very high sequence homology to the Alu-J repetitive element found in the human genome [76] and the Alu domain of 7SL RNA (Fig. 1B) [25,76]. In terms of evolution, 7SL RNA is in fact the progenitor of BC200, and BC200 is the result of retroposition of Alu monomers, followed by exaptation into the human genome [79]. Importantly, the Alu region of both BC200 and 7SL genes contains A box and B box internal Pol III promoter elements for efficient transcription [25,80]. This region also contains the binding site for the SRP protein heterodimer, SRP9/14, as discussed below [81].
2.2.3. Analogues of BC200
Since the evolutionary event that led to the emergence of BC200 is relatively recent, no orthologs of BC200 have been identified outside of the primate order [37]. However, BC200 shares characteristics with lncRNAs in other species, suggesting that they could share similar functions in the cell [37,82].
Brain cytoplasmic 1 (BC1) is a non-coding RNA of 152 nucleotides that is specific to rodents [83,84]. In addition to sharing the same tissue and cellular localisation as BC200 in neuronal dendrites [25,84], BC1 is characterised by a tripartite structure, similar to BC200, comprised of a 5′ domain that forms an extended stem loop structure, a central A-rich region and a 3′ terminal stem loop structure [85]. This reflects its evolutionary origin by retroposition of alanine transfer RNA (tRNAAla) [83]. Moreover, BC1 is transcribed by Pol III [39] and appears to play a role in translational regulation [86]. Indeed, BC1 and BC200, despite their independent evolutionary origins, have very similar sequences (Fig. 1B); therefore, it is not surprising that both of these non-coding RNAs interact with some of the same cellular proteins (Section 3).
Potential analogues to BC200 have also been found outside of mammals in Caenorhabditis elegans (C. elegans), although the C. elegans non-coding transcriptome is not fully characterised. A recent RNA sequencing study identified three potential analogues of BC200, based on the observation that the 5′ region of the non-coding RNAs inc394, inc465 and inc467 show high sequence similarity with the BC200 5′ Alu domain [87]. Similar to BC200 and BC1, each of the C. elegans transcripts contains an internal promoter element, approximately 10–20 nucleotides downstream of the transcription start site, suggesting that these RNAs could be transcribed by Pol III [87,88] Further sequence analysis showed that inc394 shares the greatest similarity to BC200, with a sequence identity of up to 65% when sequence gaps are excluded [87]. In terms of functional analysis, inc394, inc465 and inc467, were shown to interact with the orthologous RNA binding complex, SRP9/14 [65,81,87]. While further characterisation of C. elegans non-coding RNAs is needed, it certainly raises the possibility that BC200 may have other analogues that will be uncovered with RNA sequencing approaches.
3. BC200 protein binding partners: a reflection of its function?
Since BC200, BC1 and 7SL RNAs share many sequence characteristics (Fig. 1C), it is plausible to assume that they could have overlapping functions in the cell through their interactions with proteins.
Here we highlight currently known BC200 protein binding partners and indicate that many proteins have been shown to also interact with BC1 or 7SL RNAs. By analysing these interactions and determining how these lncRNA-protein complexes affect cell function, future studies can apply this information to determine the role(s) that BC200 plays in cancer (Table 1).
Table 1.
BC200 protein interactors. Some protein interactors are common between BC200, BC1 and 7SL RNAs; whereas others interact only with BC200. This suggests that the RNAs, along with their associated proteins, form different functional complexes in cells.
| Protein name | Protein function | Role of the complex with BC200 | Experimental system | Reference | |
|---|---|---|---|---|---|
| Common to BC200 and BC1 | La | Human autoantigen; many aspects of RNA metabolism | Links BC200 to the ribosome | Human and rat brain lysate | [63] |
| FMRP | mRNP transport and translation | Stabilizes interaction of BC200 with mRNA | Mouse brain lysate | [68] | |
| PABP | Regulates translation initiation | BC200 sequesters PABP, preventing translation initiation of other polyadenylated mRNAs | Human and mouse brain lysate | [66,102] | |
| Purα | RNA binding protein | BC200 is a linker between the protein and mRNA | In vitro system | [104] | |
| eIF4A | Helicase | Prevents formation of 48 S complex | Rat brain lysate | [86] | |
| eIF4B | Controls translation initiation | BC200 blocks eIF4B to prevent translation initiation | In vitro system | [60] | |
| Common to BC200 and 7SL | SRP9/14 | Translational elongation arrest | Unknown | Primate brain lysate | [65,81] |
| Interaction only with BC200 | SYNCRIP | Alternative splicing regulation, polyadenylation, mRNA metabolism, transport | Multifunctional, ultimately involved in synaptic plasticity | Human brain lysate | [64,112] |
| RHAU/DX36 | Helicase | BC200 stabilizes unwound RNA | HEK293T, MCF-7, HeLa, SK-BR-3 cells | [118] | |
| hnRNP E1/E2 | Translation activation | Translation regulation | Rabbit reticulocyte lysate | [120] | |
| hnRNP A2/B1 | Alternative splicing regulation | Splicing of Bcl-x | MCF7 cells | [28] |
3.1. BC200 and BC1 share protein partners
This section summarises information regarding a number of proteins that bind to both BC200 and BC1. Considering the neuronal expression of BC200 and BC1 [25,84], many of these studies focus specifically on interactions in the dendrites of neurons.
3.1.1. La protein
La protein is found in nearly all eurkaryotic cells and is a human auto-antigen that binds to terminal poly-uridylate tails (UUU-3′OH) of Pol III transcripts [89,90]. It is involved in many different aspects of RNA metabolism and regulates the downstream processing of RNAs, including transport and cap-independent translation [91,92]. In their study, Kremerskothen et al. showed in vitro and in vivo that BC1 and BC200 interacts with La, and that the interaction with BC200 was dependent on the 3′ end of the transcript [63]. From this study, the authors propose that a La homodimer bridges the 5′ Alu domain of BC200 to the ribosome to exert translational elogation arrest, together with SRP9/14 [63]. Another study, however, suggests that the interaction with BC200 and La is not part of a functional RNP complex in neuronal dendrites because the La protein cannot be detected [66].
3.1.2. Fragile X mental retardation protein (FMRP)
FMRP is a neuronal protein involved with messenger ribonucleoprotein (mRNP) transport and localised protein synthesis at synapses, which is critically important for synaptic plasticity [[93], [94], [95]]. Mutated or absent FMRP is responsible for the fragile X mental retardation syndrome, the most common cause of inherited mental retardation [94]. While FMRP is involved in mRNP transport and translational repression of mRNAs, it has also been shown to interact with BC200 and BC1 [68,96]. In their work, Zalfa and colleagues propose that the complex formed by FMRP and BC RNAs stabilizes or facilitates the interactions with FMRP-targeted mRNAs that are translationally repressed. In addition, they hypothesize that polyA-binding protein (PABP) might further stabilize this complex [96].
Interactions between BC200/BC1 and FMRP are not clear cut however. Kremerskothen et al. (1998) reported conflicting data, indicating that BC200 does not bind to FMRP [63]. While others report that FMRP and BC1 are unable to form a specific complex in vitro under physiological conditions [97]. Additionally in 2008, Iacoangeli et al. demonstrated that FMRP function in translational inhibition was independent of BC1 [98]. Moreover, they did not find evidence of an interaction between BC1 and FMRP in vitro or in vivo and show that FMRP is binding to mRNA targets in BC1 knockout mice, similar to that observed in wild-type animals [98]. Indeed, a subsequent study suggested that FMRP and BC1 RNA act in a sequential, but independent way to inhibit activity-stimulated translation [99]. Despite the efforts of many groups, the precise mechanisms of FMRP function through any engagement with BC200 lncRNA are still unclear.
3.1.3. Poly A-Binding protein (PABP)
PABP is an RNA-binding protein that binds mostly to the poly(A) tails of mRNA [100]. As both BC200 and BC1 non-coding RNAs have A-rich central domains (Fig. 1B), Muddasheety et al. investigated the possibility that PABP is associated with BC200/BC1 RNPs [66]. Since PABP influences translation initiation through an interaction with eukaryotic initiation factor 4G (eIF4G) [101], the authors proposed that BC200 and BC1 bind to PABP, forming stable RNPs that can modulate translation initiation [66]. Alternatively, Kondrashov et al. (2005) suggested that BC200 exerts its translational inhibitory effects by acting as a competitor for PABP [102]. This is supported by a subsequent study that observed strong inhibitory effects of naked BC200/BC1 RNAs on translation in reticulocyte lysate and transfected cell systems, while the inclusion of PABP reduced this effect [61]. One conclusion from these studies is that other protein partners may also play a role in BC200-mediated translational repression. Such an example is eIF4A, which will be discussed below. Indeed, Lin et al. attribute less than 20% of translational repression competence to the sequestration of PABP by BC200 [62].
3.1.4. Purα
Purα is a single-stranded DNA and RNA-binding protein that is conserved throughout evolution and has multiple roles, from transcriptional activation to cell growth [103]. Using a knockout mouse system, it was demonstrated that Purα has an essential role in dendrite development [104]. As dendrites contain numerous localised mRNAs [105] and BC200/BC1 [25,84], Johnson et al. (2006) were the first to link Purα binding to an mRNA known to be translated in dendrites, microtubule-associated protein 1B mRNA (Map1B), and to BC200 and BC1 non-coding RNAs [104]. Interestingly, sequence complementarity between Map1B mRNA and BC200/BC1 RNAs had been noted in an earlier study [96]. Combining these observations, Johnson et al. proposed that Purα functions with BC200/BC1 to target certain mRNAs for localised translation in dendrites. This supports the hypothesis that BC200 may act as a linker between certain proteins and mRNAs, allowing transient regulation and/or modification. Indeed the possibility of BC200 interactions with other RNAs has not been fully explored, particularly outside of a neuronal context.
3.1.5. Eukaryotic initiation factor 4A (eIF4A) and eukaryotic initiation factor 4B (eIF4B)
Eukaryotic initiation factors are essential for translation initiation, including events such as cap-recognition and binding of the 48 S pre-initiation complex to mRNAs [106]. To examine the role of BC200/BC1 RNAs in this process, Wang and al. showed that BC1 is capable of inhibiting the formation of the 48 S initiation complex by targeting eIF4A, an ATP-dependent, RNA helicase [86]. In 2008, Lin et al. went on to show that BC200 and BC1 interfere with eIF4A's catalytic mechanism by blocking the factor's helicase activity, while enhancing its ATPase activity [62]. Both studies suggest a role in translational repression for these two non-coding RNAs.
Continuing with BC200's effects on eurkaryotic translation initiation factors, another report shows that neuronal BC RNAs mediate translational inhibition by binding to eIF4B. In doing so, BC200/BC1 prevents this factor from interacting with 18 S ribosomal RNA (rRNA) in the small ribosomal subunit [59]. In this case, the BC RNAs are acting as competitors of 18 S rRNA for access to eIF4B. In an extension of this work, a subsequent study demonstrated that BC RNA interactions with eIF4B decrease upon neural stimulation that results in dephosphorylation of eIF4B [60]. Once the BC RNAs have reduced affinity for dephosphorylated eIF4B, the factor can fully participate in translation initiation [60]. Significantly, this work highlights the possibility of post-translational modifications as a level of regulation in BC200-protein complex formation and activity.
3.2. BC200 and 7SL – Alu-interacting heterodimer, signal recognition particle proteins 9 and 14 (SRP9/14)
Several studies have shown that SRP9/14 and BC200 interact [61,65,81]. SRP9/14 heterodimer is a well-known component of the signal recognition particle (SRP), an RNP that recognises and co-translationally targets secretory proteins to the endoplasmic reticulum (ER) [107]. As mentioned earlier, 7SL RNA is a 300-nucleotide, non-coding RNA that forms the scaffold of SRP [78] and interacts with six polypeptides, SRP9/14, SRP19, SRP54, SRP68 and SRP72 [108]. In SRP, the heterodimer SRP9/14 mediates translation elongation arrest, pausing translation briefly to allow engagement between the signal sequence of the nascent polypeptide and the translocon machinery in the ER membrane [109,110].
Given the sequence conservation between BC200 and 7SL, Bovia et al. were the first to show that human SRP9/14 interacts with BC200 lncRNA; this result was subsequently confirmed in vivo by another group [65]. Unsurprisingly, rodent SRP9/14 does not interact with BC1, which does not have an Alu domain [81]; however, a recent study on BC200 analogues in C.elegans shows that inc394, inc465 and inc467 do interact with C.elegans SRP9/14 [87]. It remains to be assessed whether this common characteristic between humans and C.elegans has functional meaning.
Despite SRP9/14 and BC200's involvement in translational events, the specific role of the interaction between the heterodimer and BC200, as components of an RNP complex, still remains unclear.
3.3. BC200-specific protein interactors
3.3.1. Synaptotagmin-binding, cytoplasmic, RNA-interacting protein (SYNCRIP)
SYNCRIP [64] is part of the heterogeneous nuclear ribonucleoprotein (hnRNP) protein family that is involved in splicing regulation, polyadenylation and other aspects of mRNA metabolism and transport [111]. The interaction between BC200 and SYNCRIP was first described in a study where the authors showed that the A-rich region of BC200 binds to SYNCRIP's N-terminal RNA recognition motifs (RRMs) in vitro binding assays, combined with immunoprecipitation from human brain extracts [64]. SYNCRIP had previously been described as a component of RNA granules and was implicated in RNA localisation and/or translational control in dendrites [[112], [113], [114]]. Therefore, SYNCRIP could support BC200 translational repressor function by playing the role of a linker between neuronal RNA granules and the dendritic translation machinery [64].
As Duning et al. (2008) solely investigated BC200/SYNCRIP interaction in a neuronal context, it would be of interest to determine if a similar interaction exists in cancer cells. One possible lead regarding a potential link to cancer comes from Grosset et al. (2000), who discovered that the mRNA of the proto-oncogene, c-fos, is stabilised by a protein complex that includes NSAP1, also known as SYNCRIP [115]. Furthermore, this study discussed the role of SYNCRIP as a bridge between the major protein-coding region determinant of instability and the poly(A) tail of c-fos, thereby preventing its deadenylation and subsequent degradation [115]. Given the ability of BC200 to bind to both SYNCRIP and PABP, it is interesting to speculate whether the lncRNA might act as a scaffold to facilitate the interaction.
3.3.2. RNA helicase associated with AU-rich element (RHAU/DHX36)
RHAU/DHX36 is an ATP-dependent, DEAH-box, RNA helicase that can unwind both DNA and RNA quadruplexes [116,117]. By performing RNA co-immunoprecipitation screens to identify novel RNAs that interact with RHAU, Booy and al. (2015) identified BC200 [118]. Contrary to the repressive control BC200 imposes on eIF4A helicase activity, the BC200-RHAU interactions did not appear to impair RHAU helicase function; however, they did characterise a RHAU-dependent ability of BC200 to bind to unwound RNA quadruplex sequences [118]. From their work, the authors postulated that RHAU may recruit BC200 to stabilise and prevent refolding of unwound RNA quadruplexes [118], providing further evidence that BC200's cellular roles will likely involve interactions with proteins and other RNAs.
3.3.3. Heterogeneous nuclear ribonucleoprotein E1 and E2 (hnRNP E1 and E2)
Besides SYNCRIP descibed above, other hnRNP family members also interact with BC200. Using yeast three-hybrid screening [119], Jang et al. (2016) identified two novel, candidate binding partners of BC200, hnRNP E1 and E2, that were subsequently confirmed by electromobility shift assays [120]. Given the role of BC200 in translational repression, the group investigated the link between these new interactors and this function. They found that translation of a luciferase reporter gene was inhibited by BC200; however, the addition of E1 and E2 relieved the inhibition [120]. Since this was an in vitro investigation, future work will likely examine whether the BC200 interacts with hnRNP E1 and E2 in vivo.
3.3.4. Heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP A2/B1)
Although nearly all of the previous studies suggest that BC200 function is related to translational repression, a recent study has reported that BC200 is involved in splicing through an interaction with the RNA-binding protein hnRNPA2/B1 in breast cancer cells [28]. Singh et al. (2016) sought to understand the mechanism of BC200, following the observation that BC200 is highly expressed in breast cancer cells [28]. To do this, they created a CRISPR/Cas9 generated, BC200 knockout cell line. Using the BC200 knockout cell line, they observed a suppression of cell growth, due to expression of Bcl-xS, the pro-apoptotic isoform of Bcl-x [121]. Through subsequent experiments, the authors demonstrate that BC200 interacts with Bcl-x mRNA and hnRNP A2/A1. These interactions inhibit the splicing factor, Sam68, thereby promoting formation of the short, pro-apoptotic protein, Bcl-xS. In this scenario, BC200 plays an oncogenic role in breast cancer.
In conclusion, while some partners of BC200 are common with those of BC1 or 7SL RNAs, other studies have found specific protein interactors to BC200. While many of the identified BC200-binding proteins support a function for BC200 in translation repression, it is likely that BC200 RNPs are dynamic. With one study demonstrating that BC200 is involved in the regulation of the alternative splicing in cancer cells, this raises the question as to whether this function only appears in a disease context due to upregulation of BC200 expression. Based on the evidence so far, we suspect that BC200 is necessary at multiple regulatory levels, e.g. in transport of mRNA to the dendrite and followed by regulation of mRNA translation in the dendrite. It is quite possible that BC200 and its binding partners form RNA regulons to coordinate multiple cellular events [122].
4. BC200 RNA in disease contexts
Although lncRNAs are usually expressed at lower levels than protein-coding genes in physiological contexts, many lncRNAs have dysregulated expression in pathological contexts, such as neurodegenerative disorders or cancer. BC200 is not different in this regard (Table 2), and here we focus on our current understanding of BC200 expression in neurological disease and cancer and the utility of BC200 in a clinical context (Fig. 2).
Table 2.
BC200 lncRNA expression in disease contexts. BC200 RNA levels vary across disease contexts. However, in many cancer types, studies indicate the BC200 is present at higher levels.
| Pathology | Tissue | BC200 expression associated | Reference |
|---|---|---|---|
| Alzheimer's Disease | Brain | Up/Down | [[124], [125], [126], [127]] |
| Cancer | Breast | Up | [28,31,70,71,[147], [148], [149]] |
| Cervix | Up | [33] | |
| Ovary | Up/Down | [29,70] | |
| Lung | Up | [27,70] | |
| Parotid | Up | [70] | |
| Tongue | Up | [70] | |
| Oesophagus | Up | [70,150] | |
| Stomach | Up | [32] | |
| Colon | Up | [34] |
Fig. 2.
Expression of BC200 in association with disease contexts.
BC200 lncRNA has been linked to many human diseases, mostly through observed alterations to its RNA expression levels in normal versus affected tissue or tumour (Table 2). To fully understand how BC200 can affect cells and influence diseases, detailed examination of its biological roles in each disease context is required. This will require biochemical characterisation of BC200's molecular interactions and experimental determination of how these interactions directly influence cellular processes.
4.1. BC200/BC1 RNA in neurodegenerative disease
4.1.1. BC200 is altered in Alzheimer's disease (AD)
BC200 has been implicated in Alzheimer disease (AD) due to its altered expression [123,124]. Mus et al. (2007) found that in normal aging, levels of BC200 are reduced by more than 60% between the ages of 49 and 86 [125]. In comparison, BC200 RNA levels were significantly elevated in AD brain, compared to normal matched controls [125]. Since BC200 RNA was specifically observed in brain regions that are related to the disease, it was hypothesized that elevated BC200 RNA is involved with synaptodendritic deterioration in AD neurons [125]. However, this contradicts an earlier report that stated that AD-affected neocortex exhibits a 70% reduction in BC200 levels compared to age-matched controls [126]. While the difference might be explained by sampling location [123], BC200 appears to be dysregulated in AD [124]. Some studies have tried to understand the role of BC200 in AD, questioning whether it affects microtubule-dependent mRNP transport, contributing to axonal and dendritic blockage [124]. However, another study has linked BC200 with necrosis [127].
4.1.2. BC200 rodent analogue, BC1, is implicated in the regulation of neuronal receptors
At present, literature detailing the role of BC200 in neurological pathologies remains confined to AD. However, the rodent analogue BC1 has been documented to mediate signalling events within neurons. In a study lead by Centonze [128], BC1 knockout mice were used to assess the influence of this RNA on the dopamine D2 receptor (D2DR). D2DR is a G-protein coupled receptor found in dopaminergic neurons that upon stimulation initiates a range of signalling cascades that mediate various physiological responses [129]. Interestingly, the study by Centonze et al. [128] determined that BC1 is a regulator of dopamine transmission. The group concluded that the mechanism, though not fully defined, is not the result of direct translational regulation of the receptor. Instead, BC1 seemed to affect proteins involved in the receptor's turnover and/or stability [128]. In another BC1 study, Zhong and co-workers [130] demonstrated that BC1 regulates group 1 metabotropic glutamate receptors (mGluRs) and is necessary for excitation-repression homeostasis in neurons. The absence of BC1 in vivo animal models was shown to provoke epileptogenic vulnerability, indicative of neuronal hyperexcitability [130]. Importantly both studies link BC1 regulatory RNA with neuronal receptor function and synaptic plasticity, which impacts overall brain function [93,131]. In humans, D2DR and mGluRs are implicated, and to some degree targeted therapeutically, in neurodegenerative diseases, including AD, Parkinson's (PD) and Huntingdon's (HD) [132,133].
Though BC200 shares extensive similarity with BC1 (Fig. 1C), the convergence of their functional roles, specifically in neuropathological contexts, remains to be fully explored. Despite the scientific investigations so far, the exact role of BC200 in AD and other neurodegenerative diseases is unclear, with further studies necessary.
4.2. BC200 in cancer
4.2.1. Association of BC200 with cancer
Since the discovery of the existence of lncRNAs, numerous studies have focused on identification, differential expression, and to a more limited degree, the underlying biological roles of lncRNAs in cancer. There are many excellent reviews on this topic [3,134,135]. Without doubt, the fast pace at which lncRNAs are being identified through RNA sequencing efforts is continuing [2,16,136]. While we have collated information about BC200's association with cancer below, there are still many unanswered questions regarding how the lncRNA specifically influences the altered biology of a cancer cell. Given BC200's many unique characteristics, i.e. Pol III transcript, mostly cytoplasmic localisation and small size, it may be that BC200 will occupy different functional roles, compared to more well-characterised, cancer-associated lncRNAs, such as MALAT1 and HOTAIR, that have nuclear roles in regulation of gene expression [137,138].
The initial suggestion that BC200 might have any role in cancer was described ten years ago. Fundamental work by Chen et al. [70] demonstrated that BC200 lncRNA was detectable in carcinomas of the breast, cervix, oesophagus, lung, ovary, parotid and tongue, while no expression was found in normal tissue, through immunohistochemistry analyses of tissue sections. The authors were the first to suggest a link between BC200 and the induction and/or progression of the tumour, but there was no molecular evidence to support that association directly [70].
While upregulation of BC200 RNA expression in cancer cells is not reflective of an overall increase in Pol III transcription [70], there is a report that BC200 expression is responsive to c-MYC and that c-MYC binds to the BC200 promoter region in non-small-cell lung cancer (NSCL) cells [27]. Given many cancers are driven by the MYC oncogene [42,43], it is quite possible that BC200 expression will be higher in MYC-driven cancers, although this has not been exhaustively examined. Interestingly a new report found that elevated Pol III transcription in early breast cancer tumourigenesis is driven by epigenetic changes, coupled with MYC [139]. Epigenetic control of BC200 expression is currently unknown.
4.2.2. Role of BC200 in cancer
Although the association between BC200 and cancer is established, numerous questions remain: how is BC200 expressed in cancer contexts, when it cannot be detected in normal tissue? Is the unusual BC200 pattern of expression functionally relevant with the cancer progression? Are there dynamic changes to a collective set of BC200-binding proteins? Does BC200 interact with other RNAs or DNA in cancer cells? What do the molecular interactions of BC200 tell us? Are certain pathways or cell behaviours particularly affected by modulation of BC200 lncRNA levels?
Recent studies are beginning to tackle the mechanisms of BC200 action and their impact in cancer cells. In 2015, Hu and al. showed that in NSCL cancer, the oncogenic transcription factor c-MYC binds to the BC200 promoter and induces its expression [27]. Moreover, they observe that in absence of BC200, migration and invasion is affected in vitro, corresponding with a reduction in the expression of the matrix metalloproteases, MMP9 and MMP13 [27]. Secretion of these enzymes by cancer cells is necessary for the initial steps of tumour cell invasion and metastasis [140] Thus, the authors conclude that BC200 contributes to cell metastasis by promoting expression of MMP9 and MMP13 [27].
The observation that BC200 could contribute to metastasis is supported by another study that shows that after knockdown of BC200 in cervical carcinoma (HeLa) and breast cancer cell lines, cell migration is reduced [31]. However, instead of offering a mechanism involving MMPs, they propose that BC200 promotes migration and invasion by affecting the stability of the S100A11 mRNA, encoding a calcium sensing protein that is tightly associated with cell motility and invasiveness [31,141].
As described earlier, Singh et al. (2016) provided evidence of a novel regulatory function, with BC200 modulating alternative splicing of Bcl-x in the context of breast cancer [28]. Their work implies that BC200 plays an oncogenic role by negatively regulating apoptosis, allowing rogue cells to avoid cell death. In addition, their study indicated that the oestrogen receptor (ER) binds to the BC200 promoter, positively regulating its expression in breast cancer cell lines [28]. However, since most cases of advanced breast cancer have lost expression of ER [142,143], it seems likely that the expression of BC200 lncRNA may become dependent on other factors such as c-MYC as the tumour progresses [42,139,144].
In 2017 another study demonstrated that BC200 is critical for breast cancer cell survival [30]. They found that cell growth is inhibited, and apoptosis is induced, after knockdown of BC200 in proliferating cells [30]. Leading on from this observation, this raises the possibility of BC200 as a candidate for specific drug targeting of tumour cells. The same study notes that the lethality of BC200 knockdown is restricted to actively proliferating cells [30], making BC200 an attractive cancer therapeutic target.
Adding to the existing literature, there have been a few studies published in 2018 indicating that BC200 is overexpressed in colon, stomach and cervical cancers [[32], [33], [34]]. In each of these studies, BC200 RNA levels were elevated in tumour versus adjacent normal tissues from patients, as determined by qRT-PCR. To begin to characterise how BC200 influences cancer cell behaviour, Ren et al. [32], Peng et al. [33] and Wu et al. [34] utilized corresponding cell lines to modulate BC200 expression by siRNA knockdown. Interestingly, each study revealed different possible mechanisms of BC200 action (Table 3). These included upregulation of EpCAM [32]; targeting of miRNA-138 [33]; and phosphorylation of signal transducer and activator of transcription 3 (STAT3) [34]. Further experiments will certainly focus on the similarities and differences of BC200's cellular mechanisms among cancers with different tissue origins.
Table 3.
Role of BC200 in cancer progression. BC200's specific role in cancer progression remains an active area of investigation. However, some studies have begun to understand BC200's mechanism of action. Although there will likely be additional context-dependent mechanisms identified, BC200 does affect several defining features of neoplastic cells, including increased cell proliferation, migration and resistance to apoptosis.
| Cancer tissue | Process affected | Mechanism | Reference |
|---|---|---|---|
| Breast | Proliferation | Link with cell cycle progression | [28] |
| Apoptosis | Regulation of Bcl-x alternative splicing | [39] | |
| Cell migration | Modulation of S100A11 mRNA stability | [29] | |
| Cervix | Proliferation and cell migration | Targeting microRNA-138 (miR138) | [33] |
| Lung | Cell migration | Regulation of MMP9 and MMP13 expression | [27] |
| Stomach | Proliferation, apoptosis and cell migration | Regulation of EpCAM expression | [32] |
| Colon | Proliferation and cell migration | STAT3 phosphorylation; regulation of β-catenin, cyclin D1, cyclin E and c-MYC expression | [34] |
Collectively, BC200 been linked to three hallmarks of cancer [145] – cell proliferation, migration and resistance to apoptosis. Taken together, scientific findings thus far reinforce the idea that BC200 may be useful as potential therapeutic target in the treatment of multiple types of cancer.
4.2.3. Using BC200 as a cancer biomarker
As cancer research and treatment embraces new genome, transcriptome and proteome-wide technologies, there has been the emergence of “precision medicine” that heavily relies on the development and validation of biomarkers [146]. Here we highlight studies focused on the discovery of BC200 as a cancer biomarker.
In 2004, Iacoangeli et al. were the first to propose the use of BC200 as a prognostic indicator of ductal carcinoma in situ (DCIS), a non-invasive form of breast cancer [71]. They argued that BC200 is a suitable marker, as the transcript is not detected in the early stages of DCIS but gradually increases as the tumour progresses to more advanced stages of DCIS and to invasive carcinoma [71,147]. Moreover, they showed that high expression of BC200 is associated with high, nuclear-grade DCIS, which is an indicator of aggressive cancer cell behaviour [71]. Collectively, the data supported the idea that BC200 might be involved in the regulation of cellular processes that affect breast cancer migration and invasiveness [148,149]. Indeed BC200's involvement in breast cancer migration and invasiveness has been observed in subsequent studies [28,30].
Similar to studies in breast cancer, Zhao et al. showed that in oesophageal squamous cell carcinoma (ESCC), BC200 RNA expression is elevated in tumour cells compared to adjacent normal tissue [150]. Additionally they showed that high expression of BC200 RNA correlated with poor prognosis and shorter disease-free survival [150], suggesting that BC200 might be a useful prognostic marker for ESCC. Recent work by Hu et al. [27], Ren et al. [32], Peng et al. [33] and Wu et al. [34] presented similar findings of elevated BC200 RNA in tumour versus normal adjacent tissue from patient samples for lung, gastric, cervical and colon cancers. This suggests a role in tumour induction and/or progression, raising the possibility that BC200 could be clinically utilised for diagnostic and/or prognostic purposes.
Given the almost negligible expression in non-proliferating normal tissues, strong induction of BC200 may a reliable indicator of increased proliferation – one of the hallmarks of cancer [145]. Using BC200 as a biomarker may enable medical professionals and patients to make informed decisions regarding treatment, particularly given the correlation between transcript abundance and cancer progression. While this is the hope, practical use of BC200 as a cancer biomarker faces many challenges, based on the historically low success rate of clinical translation [146,151]. More realistically, it is possible that BC200 could be one of many lncRNAs to form an oncology non-coding RNA biomarker panel. This is an area that is currently undeveloped.
In summary, BC200 lncRNA has recently started to attract increased attention for its role in cancer. Although the specific details of its molecular mechanisms are currently unclear, BC200 appears to contribute to a number of different cancer hallmarks, including cell proliferation, migration and resistance to apoptosis [145]. Future work will likely reveal novel roles for BC200 lncRNA in cancer.
5. Discussion
With an increasing number of studies focusing on the role of BC200, it is the shortest, long non-coding RNA that has an emerging importance in cancer biology. With this review, we aimed to summarise previous work on BC200, while highlighting outstanding questions regarding its role and molecular interactions, particularly in cancer.
Most of the RNA-protein interactions described here are specific to the neurological context where BC200 is naturally expressed. Studies in cancer have yet to replicate these results, and there is data suggesting that BC200 function in cancer might be different to its predicted role as a translational repressor [28]. Although most studies concur that BC200 is important in several key features of cancer – like cell proliferation, survival and migration, only few studies have been able to offer a mechanism that unravels its exact function. This will certainly be the focus of future research. Indeed, with tens of thousands of different long non-coding RNAs, only a handful have been functionally characterised in cancer, like MALAT and HOTAIR [[152], [153], [154], [155]]. Therefore, a better understanding of BC200's mechanism in cancer will contribute equally to research in cancer biology and to the general knowledge on long non-coding RNAs. Simultaneously, studies focusing on BC200 as a potential diagnostic biomarker of early-stage cancers must precisely identify when its expression is detected, in addition to correlating BC200 expression with cancer development.
Acknowledgements
The authors would like to thank Mary Heapes for artwork and assistance with figures and Dr Paul Young for critical reading of the manuscript. SC is supported through the Government of Ireland Postgraduate Scholarship (GOIPG/2017/579), from the Irish Research Council. This work was supported by the Translational Research Access Programme, School of Medicine, University College Cork, Ireland.
References
- 1.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigó R., Gingeras T.R., Margulies E.H. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 4.Marchese F.P., Raimondi I., Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. doi: 10.1186/s13059-017-1348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kung J.T.Y., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilusz J.E., Freier S.M., Spector D.L. 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilusz J.E. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim. Biophys. Acta. 2016;1859:128–138. doi: 10.1016/j.bbagrm.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dieci G., Fiorino G., Castelnuovo M., Teichmann M., Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 11.White R.J. Transcription by RNA polymerase III: more complex than we thought. Nat. Rev. Genet. 2011;12:459–463. doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- 12.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hon C.-C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J.L., Gough J. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang S., Zhang L., Guo J., Niu Y., Wu Y., Li H. NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volders P.-J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2014;43:D174–D180. doi: 10.1093/nar/gku1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mercer T.R., Gerhardt D.J., Dinger M.E., Crawford J., Trapnell C., Jeddeloh J a. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol. 2011;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 2016;17:601–614. doi: 10.1038/nrg.2016.85. [DOI] [PubMed] [Google Scholar]
- 19.Amaral P.P., Mattick J.S. Noncoding RNA in development. Mamm. Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 20.Rashid F., Shah A., Shan G. Long non-coding RNAs in the cytoplasm. Dev. Reprod. Biol. 2016;14:73–80. doi: 10.1016/j.gpb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dykes I.M., Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Dev. Reprod. Biol. 2017;15:177–186. doi: 10.1016/j.gpb.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long Y., Wang X., Youmans D.T., Cech T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017;3 doi: 10.1126/sciadv.aao2110. eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watson J.B., Sutcliffe J.G. Primate brain-specific cytoplasmic transcript of the Alu repeat family. Mol. Cell Biol. 1987;7:3324–3327. doi: 10.1128/mcb.7.9.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng G., Tiedge H., Brosius J. Expression of dendritic BC200 RNA, component of a 11.4S ribonucleoprotein particle, is conserved in humans and simians. Neurosci. Lett. 1997;224:206–210. doi: 10.1016/s0304-3940(97)13471-1. [DOI] [PubMed] [Google Scholar]
- 25.Tiedge H., Chen W., Brosius J. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J. Neurosci. 1993;13 doi: 10.1523/JNEUROSCI.13-06-02382.1993. 2382 LP-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W., Böcker W., Brosius J., Tiedge H. Expression of neural BC200 RNA in human tumours. J. Pathol. 1997;183:345–351. doi: 10.1002/(SICI)1096-9896(199711)183:3<345::AID-PATH930>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 27.Hu T., Lu Y.-R.R. BCYRN1, a c-MYC-activated long non-coding RNA, regulates cell metastasis of non-small-cell lung cancer. Canc. Cell Int. 2015;15:36. doi: 10.1186/s12935-015-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh R., Gupta S.C., Peng W.-X.X., Zhou N., Pochampally R., Atfi A. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.168. e2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu D., Wang T., Ren C., Liu L., Kong D., Jin X. Downregulation of BC200 in ovarian cancer contributes to cancer cell proliferation and chemoresistance to carboplatin. Oncol. Lett. 2016;11:1189–1194. doi: 10.3892/ol.2015.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Booy E.P., McRae E.K.S., Koul A., Lin F., McKenna S.A. The long non-coding RNA BC200 (BCYRN1) is critical for cancer cell survival and proliferation. Mol. Canc. 2017;16:1–15. doi: 10.1186/s12943-017-0679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin H., Lee J., Kim Y., Jang S., Lee Y.Y., Kim S. Knockdown of BC200 RNA expression reduces cell migration and invasion by destabilizing mRNA for calcium-binding protein S100A11. RNA Biol. 2017 doi: 10.1080/15476286.2017.1297913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren H., Yang X., Yang Y., Zhang X., Zhao R., Wei R. Upregulation of LncRNA BCYRN1 promotes tumor progression and enhances EpCAM expression in gastric carcinoma. Oncotarget. 2018;9:4851–4861. doi: 10.18632/oncotarget.23585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng J., Hou F., Feng J., Xu S.-X., Meng X.-Y. Long non-coding RNA BCYRN1 promotes the proliferation and metastasis of cervical cancer via targeting microRNA-138in vitroandin vivo. Oncol. Lett. 2018;15:5809–5818. doi: 10.3892/ol.2018.8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu K., Xu K., Liu K., Huang J., Chen J., Zhang J. Long noncoding RNA BC200 regulates cell growth and invasion in colon cancer. Int. J. Biochem. Cell Biol. 2018;99:219–225. doi: 10.1016/j.biocel.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Basile V., Vicente A., Martignetti J.A., Skryabin B.V., Brosius J., Kennedy J.L. Assignment of the human BC200 RNA gene (BCYRN1) to chromosome 2p16 by radiation hybrid mapping. Cytogenet. Cell Genet. 1998;82:271–272. doi: 10.1159/000015117. [DOI] [PubMed] [Google Scholar]
- 36.Ludwig A., Rozhdestvensky T.S., Kuryshev V.Y., Schmitz J., Brosius J. An unusual primate locus that attracted two independent Alu insertions and facilitates their transcription. J. Mol. Biol. 2005;350:200–214. doi: 10.1016/j.jmb.2005.03.058. [DOI] [PubMed] [Google Scholar]
- 37.Martignetti J.A., Brosius J. BC200 RNA : a neural RNA polymerase III product encoded by a monomeric Alu element. Proc. Natl. Acad. Sci. Unit. States Am. 1993;90:11563–11567. doi: 10.1073/pnas.90.24.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y., Lee J., Shin H., Jang S., Kim S.C., Lee Y. Biosynthesis of brain cytoplasmic 200 RNA. Sci. Rep. 2017;7:6884. doi: 10.1038/s41598-017-05097-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martignetti J.A., Brosius J. BC1 RNA: transcriptional analysis of a neural cell-specific RNA polymerase III transcript. Mol. Cell Biol. 1995;15:1642–1650. doi: 10.1128/mcb.15.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orioli A., Pascali C., Pagano A., Teichmann M., Dieci G. RNA polymerase III transcription control elements: themes and variations. Gene. 2012;493:185–194. doi: 10.1016/j.gene.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 41.Vannini A., Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 42.Dang C.V. 2012. MYC on the Path to Cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalkat M., De Melo J., Hickman K., Lourenco C., Redel C., Resetca D. MYC deregulation in primary human cancers. Genes (Basel) 2017;8:151. doi: 10.3390/genes8060151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yates A., Akanni W., Amode M.R., Barrell D., Billis K., Carvalho-Silva D. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–D716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin H., Lee J., Kim Y., Jang S., Ohn T., Lee Y. Vol. 50. 2017. pp. 318–322. (Identifying the Cellular Location of Brain Cytoplasmic 200 RNA Using an RNA-recognizing Antibody). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ingolia N.T., Brar G.A., Stern-Ginossar N., Harris M.S., Talhouarne G.J.S., Jackson S.E. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Heesch S., van Iterson M., Jacobi J., Boymans S., Essers P.B., de Bruijn E. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014;15 doi: 10.1186/gb-2014-15-1-r6. R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carlevaro-Fita J., Rahim A., Vardy L.A., Johnson R., Guigó R., Vardy L.A. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA. 2016:1–16. doi: 10.1261/rna.053561.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson D.M., Anderson K.M., Chang C.L., Makarewich C.A., Nelson B.R., McAnally J.R. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guttman M., Russell P., Ingolia N.T., Weissman J.S., Lander E.S. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dinger M.E., Pang K.C., Mercer T.R., Mattick J.S. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 2008;4:1–5. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nam J., Choi S., You B., Incredible R.N.A. Dual functions of coding and noncoding. Mol. Cell. 2016;39:367–374. doi: 10.14348/molcells.2016.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rubio C.A., Weisburd B., Holderfield M., Arias C., Fang E., DeRisi J.L. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014;15:476. doi: 10.1186/s13059-014-0476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwasaki S., Floor S.N., Ingolia N.T. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature. 2016;534:558–561. doi: 10.1038/nature17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ji Z., Song R., Regev A., Struhl K. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife. 2015:4. doi: 10.7554/eLife.08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tirosh O., Cohen Y., Shitrit A., Shani O., Le-Trilling V.T.K., Trilling M. The transcription and translation landscapes during human cytomegalovirus infection reveal novel host-pathogen interactions. PLoS Pathog. 2015:11. doi: 10.1371/journal.ppat.1005288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zur H., Aviner R., Tuller T. Complementary post transcriptional regulatory information is detected by PUNCH-P and ribosome profiling. Sci. Rep. 2016;6:21635. doi: 10.1038/srep21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Michel A.M., Mullan J.P.A., Velayudhan V., O'Connor P.B.F., Donohue C.A., Baranov P.V. RiboGalaxy: a browser based platform for the alignment, analysis and visualization of ribosome profiling data. RNA Biol. 2016;13:316–319. doi: 10.1080/15476286.2016.1141862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eom T., Berardi V., Zhong J., Risuleo G., Tiedge H. Dual nature of translational control by regulatory BC RNAs. Mol. Cell Biol. 2011;31:4538–4549. doi: 10.1128/MCB.05885-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eom T., Muslimov I.A., Tsokas P., Berardi V., Zhong J., Sacktor T.C. Neuronal BC RNAs cooperate with eIF4B to mediate activity-dependent translational control. J. Cell Biol. 2014;207:237–252. doi: 10.1083/jcb.201401005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khanam T., Rozhdestvensky T.S., Bundman M., Galiveti C.R., Handel S., Sukonina V. Two primate-specific small non-protein-coding RNAs in transgenic mice: neuronal expression, subcellular localization and binding partners. Nucleic Acids Res. 2007;35:529–539. doi: 10.1093/nar/gkl1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin D., Pestova T.V., Hellen C.U.T., Tiedge H. Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol. Cell Biol. 2008;28:3008–3019. doi: 10.1128/MCB.01800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kremerskothen J., Nettermann M., op de Bekke A., Bachmann M., Brosius J. Identification of human autoantigen La/SS-B as BC1/BC200 RNA-binding protein. DNA Cell Biol. 1998;17:751–759. doi: 10.1089/dna.1998.17.751. [DOI] [PubMed] [Google Scholar]
- 64.Duning K., Buck F., Barnekow A., Kremerskothen J. SYNCRIP, a component of dendritically localized mRNPs, binds to the translation regulator BC200 RNA. J. Neurochem. 2008;105:351–359. doi: 10.1111/j.1471-4159.2007.05138.x. [DOI] [PubMed] [Google Scholar]
- 65.Kremerskothen J., Zopf D., Walter P., Cheng J.G., Nettermann M., Niewerth U. Heterodimer SRP9/14 is an integral part of the neural BC200 RNP in primate brain. Neurosci. Lett. 1998;245:123–126. doi: 10.1016/s0304-3940(98)00215-8. [DOI] [PubMed] [Google Scholar]
- 66.Muddashetty R.S., Khanam T., Kondrashov A., Bundman M., Iacoangeli A., Kremerskothen J. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. 2002;321:433–445. doi: 10.1016/s0022-2836(02)00655-1. [DOI] [PubMed] [Google Scholar]
- 67.Weinmann L., Höck J., Ivacevic T., Ohrt T., Mütze J., Schwille P. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 68.Zalfa F., Adinolfi S., Napoli I., Kühn-Hölsken E., Urlaub H., Achsel T. Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif. J. Biol. Chem. 2005;280:33403–33410. doi: 10.1074/jbc.M504286200. [DOI] [PubMed] [Google Scholar]
- 69.Sosińska P., Mikuła-Pietrasik J., Książek K. The double-edged sword of long non-coding RNA: the role of human brain-specific BC200 RNA in translational control, neurodegenerative diseases, and cancer. Mutat. Res. Mutat. Res. 2015;766:58–67. doi: 10.1016/j.mrrev.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 70.Chen W., Böcker W., Brosius J., Tiedge H. Expression of neural BC200 RNA in human tumours. J. Pathol. 1997;183:345–351. doi: 10.1002/(SICI)1096-9896(199711)183:3<345::AID-PATH930>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 71.Iacoangeli A., Lin Y., Morley E.J., Muslimov I.A., Bianchi R., Reilly J. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis. 2004;25:2125–2133. doi: 10.1093/carcin/bgh228. [DOI] [PubMed] [Google Scholar]
- 72.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 73.Menzel P., Gorodkin J., Stadler P.F. The tedious task of finding homologous noncoding RNA genes. RNA. 2009;15:2075–2082. doi: 10.1261/rna.1556009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 75.Hezroni H., Koppstein D., Schwartz M.G., Avrutin A., Bartel D.P., Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11:1110–1122. doi: 10.1016/j.celrep.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jurka J., Milosavljevic A. Reconstruction and analysis of human Alu genes. J. Mol. Evol. 1991;32:105–121. doi: 10.1007/BF02515383. [DOI] [PubMed] [Google Scholar]
- 77.Kuryshev V.Y., Skryabin B.V., Kremerskothen J., Jurka J., Brosius J. Birth of a gene: locus of neuronal BC200 snmRNA in three prosimians and human BC200 pseudogenes as archives of change in the Anthropoidea lineage. J. Mol. Biol. 2001;309:1049–1066. doi: 10.1006/jmbi.2001.4725. [DOI] [PubMed] [Google Scholar]
- 78.Walter P., Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982;299:691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- 79.Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–134. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- 80.Bredow S., Kleinert H., Benecke B.J. Sequence and factor requirements for faithful in vitro transcription of human 7SL DNA. Gene. 1990;86:217–225. doi: 10.1016/0378-1119(90)90282-v. [DOI] [PubMed] [Google Scholar]
- 81.Bovia F., Wolff N., Ryser S., Strub K. The SRP9/14 subunit of the human signal recognition particle binds to a variety of Alu-like RNAs and with higher affinity than its mouse homolog. Nucleic Acids Res. 1997;25:318–325. doi: 10.1093/nar/25.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Skryabin B.V., Kremerskothen J., Vassilacopoulou D., Disotell T.R., Kapitonov V.V., Jurka J. The BC200 RNA gene and its neural expression are conserved in anthropoidea (primates) J. Mol. Evol. 1998;47:677–685. doi: 10.1007/pl00006426. [DOI] [PubMed] [Google Scholar]
- 83.Dechiara T.M., Brosius J. Neural BC1 RNA: cDNA clones reveal nonrepetitive sequence content (rat/small RNA-derived cDNA/phage XgtlO library/identifiler elements/unique sequence oligonucleotide) Biochemistry. 1987;84:2624–2628. doi: 10.1073/pnas.84.9.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tiedge H., Fremeau R.T., Weinstock P.H., Arancio O., Brosius J. Dendritic location of neural BC1 RNA. Proc. Natl. Acad. Sci. U. S. A. 1991;88:2093–2097. doi: 10.1073/pnas.88.6.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rozhdestvensky T.S., Kopylov A.M., Brosius J., Hüttenhofer A. Neuronal BC1 RNA structure: evolutionary conversion of a tRNA(Ala) domain into an extended stem-loop structure. RNA. 2001;7:722–730. doi: 10.1017/s1355838201002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H., Iacoangeli A., Popp S., Muslimov I.A., Imataka H., Sonenberg N. Dendritic BC1 RNA: functional role in regulation of translation initiation. J. Neurosci. 2002;22:10232–10241. doi: 10.1523/JNEUROSCI.22-23-10232.2002. doi:22/23/10232 ([pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiao T., Wang Y., Luo H., Liu L., Wei G., Chen X.X.X.X. A differential sequencing-based analysis of the C. elegans noncoding transcriptome. RNA. 2012;18:626–639. doi: 10.1261/rna.030965.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Deng W., Zhu X., Skogerbø G., Zhao Y., Fu Z., Wang Y. Organization of the Caenorhabditis elegans small non-coding transcriptome: genomic features, biogenesis, and expression. Genome Res. 2006;16:20–29. doi: 10.1101/gr.4139206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wolin S.L., Cedervall T. The La protein. Annu. Rev. Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 90.Curry S., Conte M.R. A terminal affair: 3′-end recognition by the human La protein. Trends Biochem. Sci. 2006;31:303–305. doi: 10.1016/j.tibs.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 91.Maraia R.J. La protein and the trafficking of nascent RNA polymerase III transcripts. J. Cell Biol. 2001;153:13–17. doi: 10.1083/jcb.153.4.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hellen C.U., Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 93.Martin K.C., Barad M., Kandel E.R. Local protein synthesis and its role in synapse-specific plasticity. Curr. Opin. Neurobiol. 2000;10:587–592. doi: 10.1016/s0959-4388(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 94.Bagni C., Greenough W.T. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat. Rev. Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 95.Zalfa F., Achsel T., Bagni C. mRNPs, polysomes or granules: FMRP in neuronal protein synthesis. Curr. Opin. Neurobiol. 2006;16:265–269. doi: 10.1016/j.conb.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 96.Zalfa F., Giorgi M., Primerano B., Moro A., Di Penta A., Reis S. The Fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 97.Wang H., Iacoangeli A., Lin D., Williams K., Denman R.B., Hellen C.U.T. Dendritic BC1 RNA in translational control mechanisms. J. Cell Biol. 2005;171:811–821. doi: 10.1083/jcb.200506006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Iacoangeli A., Rozhdestvensky T.S., Dolzhanskaya N., Tournier B., Schutt J., Brosius J. On BC1 RNA and the fragile X mental retardation protein. Proc. Natl. Acad. Sci. Unit. States Am. 2008;105:734–739. doi: 10.1073/pnas.0710991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhong J., Chuang S.C., Bianchi R., Zhao W., Paul G., Thakkar P. Regulatory BC1 RNA and the fragile X mental retardation protein: convergent functionality in brain. PLoS One. 2010:5. doi: 10.1371/journal.pone.0015509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gorgoni B., Gray N.K. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental perspective. Briefings Funct. Genomics Proteomics. 2004;3:125–141. doi: 10.1093/bfgp/3.2.125. [DOI] [PubMed] [Google Scholar]
- 101.Tarun S.Z., Sachs A.B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 102.Kondrashov A.V., Kiefmann M., Ebnet K., Khanam T., Muddashetty R.S., Brosius J. Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP) J. Mol. Biol. 2005;353:88–103. doi: 10.1016/j.jmb.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 103.Gallia G.L., Johnson E.M., Khalili K. Puralpha: a multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res. 2000;28:3197–3205. doi: 10.1093/nar/28.17.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Johnson E.M., Kinoshita Y., Weinreb D.B., Wortman M.J., Simon R., Khalili K. Role of Pur alpha in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J. Neurosci. Res. 2006;83:929–943. doi: 10.1002/jnr.20806. [DOI] [PubMed] [Google Scholar]
- 105.Glock C., Heumüller M., Schuman E.M. mRNA transport & local translation in neurons. Curr. Opin. Neurobiol. 2017;45:169–177. doi: 10.1016/j.conb.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 106.Jackson R.J., Hellen C.U.T., Pestova T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nagai K., Oubridge C., Kuglstatter A., Menichelli E., Isel C., Jovine L. Structure, function and evolution of the signal recognition particle. EMBO J. 2003;22:3479–3485. doi: 10.1093/emboj/cdg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Walter P., Blobel G. Disassembly and reconstitution of signal recognition particle. Cell. 1983;34:525–533. doi: 10.1016/0092-8674(83)90385-9. [DOI] [PubMed] [Google Scholar]
- 109.Strub K., Walter P. Assembly of the Alu domain of the signal recognition particle (SRP): dimerization of the two protein components is required for efficient binding to SRP RNA. Mol. Cell Biol. 1990;10:777–784. doi: 10.1128/mcb.10.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Elvekrog M.M., Walter P. Dynamics of co-translational protein targeting. Curr. Opin. Chem. Biol. 2015;29:79–86. doi: 10.1016/j.cbpa.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Geuens T., Bouhy D., Timmerman V. The hnRNP family: insights into their role in health and disease. Hum. Genet. 2016;135:851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bannai H., Fukatsu K., Mizutani A., Natsume T., Iemura S.I., Ikegami T. An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J. Biol. Chem. 2004;279:53427–53434. doi: 10.1074/jbc.M409732200. [DOI] [PubMed] [Google Scholar]
- 113.Elvira G., Wasiak S., Blandford V., Tong X.-K., Serrano A., Fan X. Characterization of an RNA granule from developing brain. Mol. Cell. Proteomics. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- 114.Kanai Y., Dohmae N., Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 115.Grosset C., Chen C.Y., Xu N., Sonenberg N., Jacquemin-Sablon H., Shyu A.B. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 116.Tran H., Schilling M., Wirbelauer C., Hess D., Nagamine Y. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol. Cell. 2004;13:101–111. doi: 10.1016/s1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 117.Chen M.C., Murat P., Abecassis K., Ferré-D’Amaré A.R., Balasubramanian S. Insights into the mechanism of a G-quadruplex-unwinding DEAH-box helicase. Nucleic Acids Res. 2015;43:2223–2231. doi: 10.1093/nar/gkv051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Booy E.P., McRae E.K.S., Howard R., Deo S.R., Ariyo E.O., Dzananovic E. RNA helicase associated with AU-rich element (RHAU/DHX36) interacts with the 3’-tail of the long non-coding RNA BC200 (BCYRN1) J. Biol. Chem. 2016;291:5355–5372. doi: 10.1074/jbc.M115.711499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang B., Kraemer B., SenGupta D., Fields S., Wickens M. Yeast three-hybrid system to detect and analyze interactions between RNA and protein. Methods Enzymol. 1999;306:93–113. doi: 10.1016/s0076-6879(99)06007-3. [DOI] [PubMed] [Google Scholar]
- 120.Jang S., Shin H., Lee J., Kim Y., Bak G., Lee Y. Regulation of BC200 RNA-mediated translation inhibition by hnRNP E1 and E2. FEBS Lett. 2017;591:393–405. doi: 10.1002/1873-3468.12544. [DOI] [PubMed] [Google Scholar]
- 121.Siddiqui W.A., Ahad A., Ahsan H. The mystery of BCL2 family: Bcl-2 proteins and apoptosis: an update. Arch. Toxicol. 2015;89:289–317. doi: 10.1007/s00204-014-1448-7. [DOI] [PubMed] [Google Scholar]
- 122.Keene J.D. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 123.Wu P., Zuo X., Deng H., Liu X., Liu L., Ji A. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res. Bull. 2013;97:69–80. doi: 10.1016/j.brainresbull.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 124.Luo Q., Chen Y. Long noncoding RNAs and Alzheimer ’s disease. Clin. Interv. Aging. 2016;11:867–872. doi: 10.2147/CIA.S107037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mus E., Hof P.R., Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10679–10684. doi: 10.1073/pnas.0701532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lukiw W.J., Handley P., Wong L., Crapper McLachlan D.R. BC200 RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD) Neurochem. Res. 1992;17:591–597. doi: 10.1007/BF00968788. [DOI] [PubMed] [Google Scholar]
- 127.Liu Q., Sun S., Yu W., Jiang J., Zhuo F., Qiu G. Altered expression of long non-coding RNAs during genotoxic stress-induced cell death in human glioma cells. J. Neuro Oncol. 2015;122:283–292. doi: 10.1007/s11060-015-1718-0. [DOI] [PubMed] [Google Scholar]
- 128.Centonze D., Rossi S., Napoli I., Mercaldo V., Lacoux C., Ferrari F. The brain cytoplasmic RNA BC1 regulates dopamine D2 receptor-mediated transmission in the striatum. J. Neurosci. 2007;27:8885–8892. doi: 10.1523/JNEUROSCI.0548-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Keeler J.F., Pretsell D.O., Robbins T.W. Functional implications of dopamine D1 vs. D2 receptors: a “prepare and select” model of the striatal direct vs. indirect pathways. Neuroscience. 2014;282:156–175. doi: 10.1016/j.neuroscience.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 130.Zhong J., Chuang S.-C., Bianchi R., Zhao W., Lee H., Fenton A.A. BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J. Neurosci. 2009;29:9977–9986. doi: 10.1523/JNEUROSCI.3893-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.von Bernhardi R., Bernhardi L.E., Eugenín J. Springer; Cham: 2017. What Is Neural Plasticity? pp. 1–15. [DOI] [PubMed] [Google Scholar]
- 132.Hurley M.J., Jenner P. What has been learnt from study of dopamine receptors in Parkinson's disease? Pharmacol. Ther. 2006;111:715–728. doi: 10.1016/j.pharmthera.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 133.Ribeiro F.M., Vieira L.B., Pires R.G.W., Olmo R.P., Ferguson S.S.G. Metabotropic glutamate receptors and neurodegenerative diseases. Pharmacol. Res. 2017;115:179–191. doi: 10.1016/j.phrs.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 134.Gibb E.A., Brown C.J., Lam W.L., Stein L., Ponting C., Belgard T. The functional role of long non-coding RNA in human carcinomas. Mol. Canc. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schmitt A.M., Chang H.Y. Long noncoding RNAs in cancer pathways. Canc. Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z., Liu H. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 2014;33:306–312. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 137.Gupta R a, Shah N., Wang K.C., Kim J., Horlings H.M., David J. Long noncoding RNA HONTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Miyagawa R., Tano K., Mizuno R., Nakamura Y., Ijiri K., Rakwal R. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012;18:738–751. doi: 10.1261/rna.028639.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Park J.-L., Lee Y.-S., Song M.-J., Hong S.-H., Ahn J.-H., Seo E.-H. Epigenetic regulation of RNA polymerase III transcription in early breast tumorigenesis. Oncogene. 2017;36:6793–6804. doi: 10.1038/onc.2017.285. [DOI] [PubMed] [Google Scholar]
- 140.Pytliak M., Vargová V., Mechírová V. Matrix metalloproteinases and their role in oncogenesis: a review. Onkologie. 2012;35:49–53. doi: 10.1159/000336304. [DOI] [PubMed] [Google Scholar]
- 141.Duarte-Costa S., Castro-Ferreira R., Neves J.S., Leite-Moreira A.F. S100A1: a major player in cardiovascular performance. Physiol. Res. 2014;63:669–681. doi: 10.33549/physiolres.932712. [DOI] [PubMed] [Google Scholar]
- 142.Kuukasjärvi T., Kononen J., Helin H., Holli K., Isola J. Loss of estrogen receptor in recurrent breast cancer is associated with poor response to endocrine therapy. J. Clin. Oncol. 1996;14:2584–2589. doi: 10.1200/JCO.1996.14.9.2584. [DOI] [PubMed] [Google Scholar]
- 143.Thomas C. Gustafsson J-Å. Estrogen receptor mutations and functional consequences for breast cancer. Trends Endocrinol. Metabol. 2015;26:467–476. doi: 10.1016/j.tem.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 144.Campbell K.J., White R.J. MYC regulation of cell growth through control of transcription by RNA polymerases I and III. Cold Spring Harbor Perspect. Med. 2014;4 doi: 10.1101/cshperspect.a018408. a018408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 146.Goossens N., Nakagawa S., Sun X., Hoshida Y. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015;4:256–269. doi: 10.3978/j.issn.2218-676X.2015.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.De Leeneer K., Claes K. Non coding RNA molecules as potential biomarkers in breast cancer. Adv. Exp. Med. Biol. 2015;867:263–275. doi: 10.1007/978-94-017-7215-0_16. [DOI] [PubMed] [Google Scholar]
- 148.Perez D.S., Hoage T.R., Pritchett J.R., Ducharme-Smith A.L., Halling M.L., Ganapathiraju S.C. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum. Mol. Genet. 2008;17:642–655. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- 149.Hansji H., Leung E.Y., Baguley B.C., Finlay G.J., Askarian-Amiri M.E. Keeping abreast with long non-coding RNAs in mammary gland development and breast cancer. Front. Genet. 2014;5:1–15. doi: 10.3389/fgene.2014.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhao R.H., Zhu C.H., Li X.K., Cao W., Zong H., Cao X.G. BC200 LncRNA a potential predictive marker of poor prognosis in esophageal squamous cell carcinoma patients. OncoTargets Ther. 2016;9:2221–2226. doi: 10.2147/OTT.S99401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Poste G. Bring on the biomarkers. Nature. 2011;469:156–157. doi: 10.1038/469156a. [DOI] [PubMed] [Google Scholar]
- 152.Gutschner T., Hämmerle M., Diederichs S. MALAT1-A paradigm for long noncoding RNA function in cancer. J. Mol. Med. 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 153.Hajjari M., Salavaty A. HOTAIR : an oncogenic long non-coding RNA in different cancers. Canc. Biol. Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sun W., Yang Y., Xu C., Guo J. Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Canc. Genet. 2017;216–217:105–110. doi: 10.1016/j.cancergen.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 155.Weidle U.H., Birzele F., Gwen K., Rüger R. Long non-coding RNAs and their role in metastasis. Cancer genomics proteomics. 2017;14:143–160. doi: 10.21873/cgp.20027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kerpedjiev P., Hammer S., Hofacker I.L. Forna (force-directed RNA): simple and effective online RNA secondary structure diagrams. Bioinformatics. 2015;31:3377–3379. doi: 10.1093/bioinformatics/btv372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]