Fig. 1.
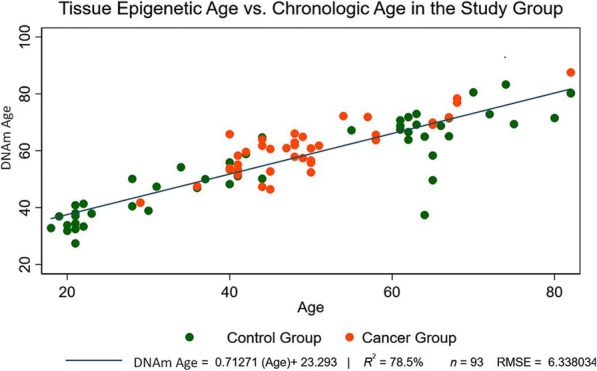
Tissue epigenetic age versus chronological age. DNA methylation age estimate based on 353 CpGs (y-axis) versus chronological age. All samples are normal breast tissue samples; normal tissue samples from cancer patients were obtained from mastectomy specimens > 3 cm from tumor margin. Samples (points) are colored by disease status of the donor: red = breast cancer and green = control. A linear regression line has been added. Age acceleration is defined as raw residual resulting from the regression model, i.e., the (signed) vertical distance to the line. Points above and below the line exhibit positive and negative epigenetic age acceleration, respectively. The high Pearson correlation coefficient r = 0.712, (p < 0.001) reflects the strong linear relationship between DNAmAge and chronological age at the time of breast sample collection
