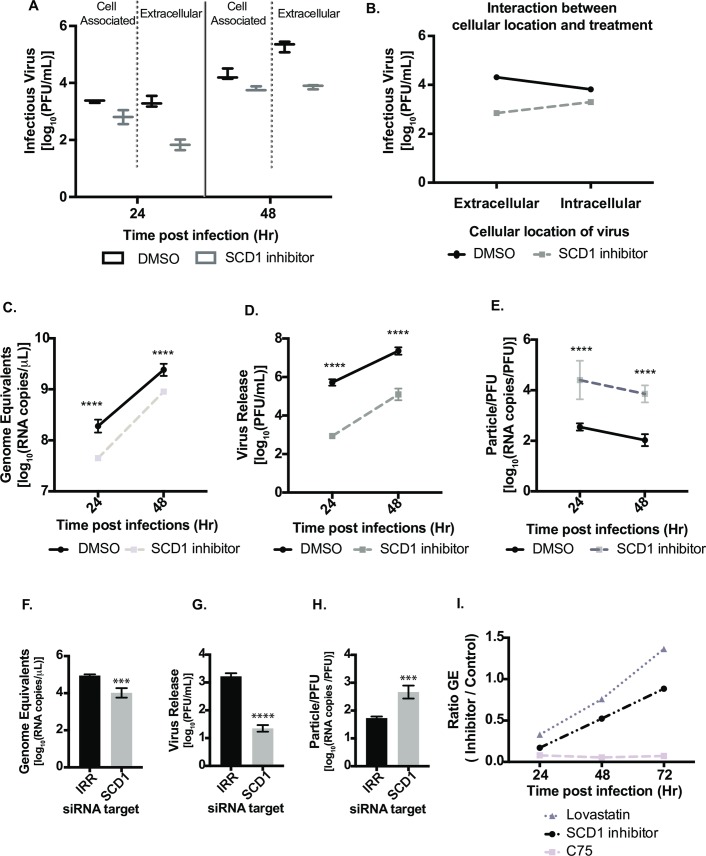
Fig 5. Inhibition of SCD1 impacts viral particle infectivity.
Huh7 cells were infected with DENV2, MOI = 1 and treated with SCD1 inhibitor or DMSO. (A) Virus in supernatants (extracellular) and cell-associated virus were collected at 24 and 48hr and quantified by plaque assay. (B) Statistical interaction plot of data from A. The non-parallel lines indicate interaction between the cellular location of the virus and the treatment. A three-way ANOVA confirmed this interaction with a p = 1.600e-06. (C-E) Huh7 cells were infected with DENV2 and treated with the SCD1 inhibitor or DMSO. Supernatants were collected at 24 and 48hr. Virus was quantified by plaque assay. Virus RNA was extracted from the same samples for qRT-PCR analysis. (C) GE were determined by qRT-PCR using a standard curve of viral RNA copies. (D) The titer of the viruses at each time point as determined by plaque assay. (E) The particle:pfu ratio was calculated by dividing the RNA copies/mL by the PFU/mL from C and D. (F-H) Huh7 cells were transfected with siRNAs for SCD1 or an irrelevant target (IRR) and incubated for 48hr. They were then infected with DENV2 (MOI = 0.1) and supernatant was collected after 24hr. Virus was quantified by plaque assay. Viral RNA was extracted from the same samples for qRT-PCR analysis. (F) GE were determined by qRT-PCR using a standard curve of viral RNA copies. (G) The titer of the viruses was determined at each time point by plaque assay. (H) The particle:pfu ratio was calculated by dividing the RNA copies/mL by the PFU/mL from F and G. (I) Comparison of GE in cells treated with the SCD1 inhibitor to those treated with lipid synthesis inhibitors, C75 and Lovastatin. [Inhibitor: (Genome equivalents/mL) / DMSO: (Genome equivalents/mL)]. (*** = p<0.005, **** = p<0.0001). GE: Genome equivalents.