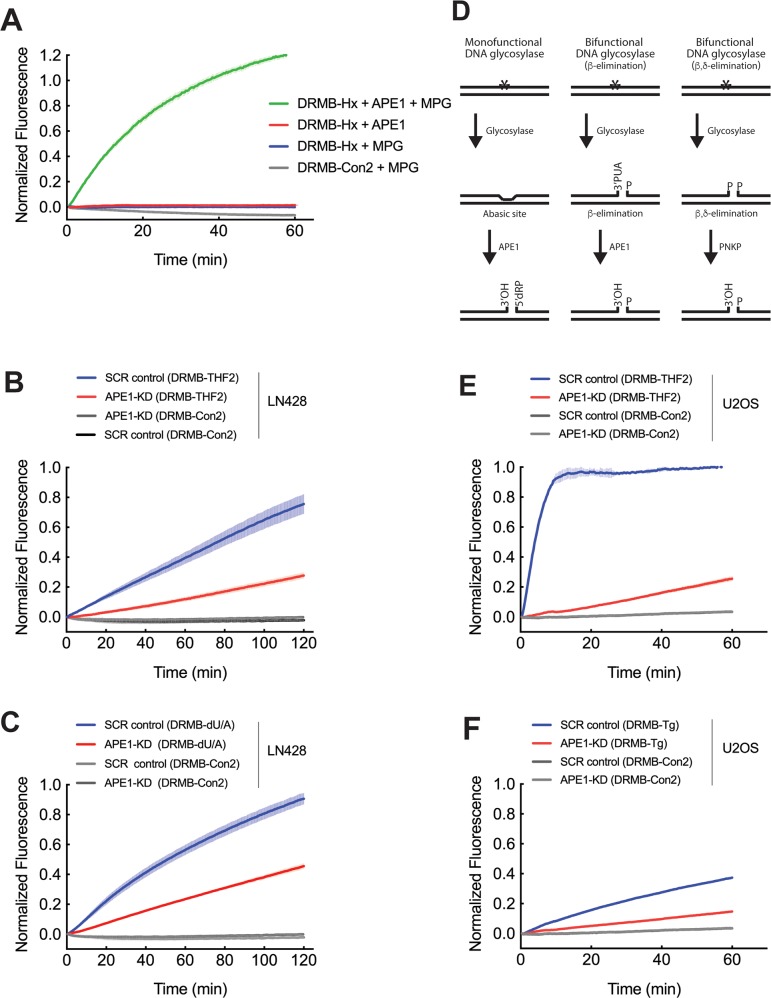
Figure 3. Both mono-functional and bi-functional DNA glycosylases depend on APE1 for DNA backbone cleavage in cell lysates.
(A) Purified methylpurine DNA glycosylase (MPG) and APE1 probed for lesion repair activity using DRMB-Hx and DRMB-Con2 assays. (B) Analysis of APE1 activity: LN429/SCR (LN429 cells expressing a scrambled control shRNA) and LN429/APE1-KD cell lysates probed for lesion repair activity using DRMB-THF2 assays, as compared to a control beacon (DRMB-Con2) containing no DNA lesion. (C) Analysis of uracil glycosylase activity: LN429/SCR (LN429 cells expressing a scrambled control shRNA) and LN429/APE1-KD cell lysates probed for lesion repair activity using DRMB-dU/A assays, as compared to a control beacon (DRMB-Con2) containing no DNA lesion. (D) Simplified schematic detailing the lesion removal and strand cleavage activities of mono-functional and bi-functional DNA glycosylases and APE1 in BER. Mono-functional DNA glycosylase activity (Left) leaves an abasic site as a substrate for APE1, resulting in DNA strand cleavage. Bifunctional DNA glycosylases excise the base and then catalyze either (Center) β-elimination, resulting in a 3’phospho, unsaturated aldehyde (PUA) and a 5’phosphate (P) in the gap that is then further processed by APE1. (Right) β,δ-elimination resulting in a 3’phosphate (P) and a 5’phosphate (P) in the gap that can then be further processed by polynucleotide kinase 3′-phosphatase (PNKP). (E) Analysis of APE1 activity: U2OS/SCR (U2OS cells expressing a scrambled control shRNA) and U2OS/APE1-KD cell lysates probed for lesion repair activity using DRMB-THF2 assays, as compared to a control beacon (DRMB-Con2) containing no DNA lesion. (F) Analysis of glycosylase activity for removal of thymine glycol: U2OS/SCR (U2OS cells expressing a scrambled control shRNA) and U2OS/APE1-KD cell lysates probed for lesion repair activity using DRMB-Tg assays, as compared to a control beacon (DRMB-Con2) containing no DNA lesion. Plot data show normalized fluorescence values and are the mean of three independent experiments with error bars representing SEM. Statistical values are listed in Supplementary Tables 1 and 2.