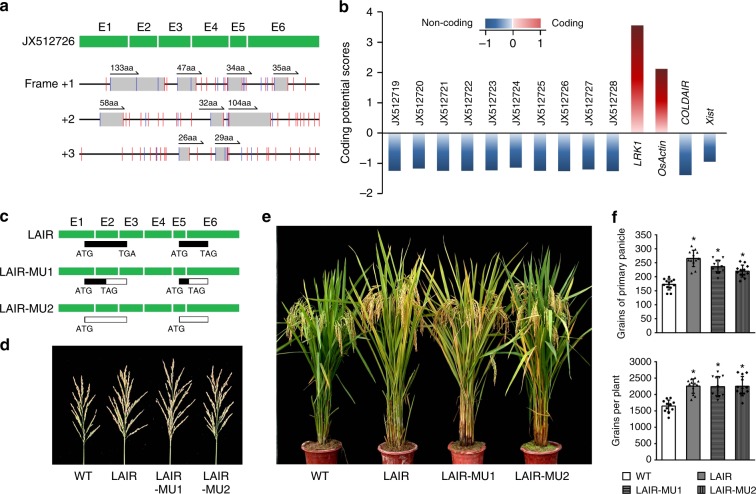
Fig. 2.
LAIR coding potential analysis. a Analysis of the LAIR open reading frame (JX512726). All frames (gray boxes) were identified in the three forward frames. The two longest open reading frames encoded 133 and 104 amino acids (aa). Blue and red lines represent the start and stop codons, respectively. b Analysis of the coding potential of the alternatively spliced LAIR isoforms (Supplementary Fig. 2). Coding potential scores were generated using the CPC program. Transcripts with scores beyond −1 and 1 are marked as non-coding or coding in this CPC classification28. LRK1 and OsActin are provided as coding examples, while COLDAIR and Xist represent non-coding examples. c LAIR-mutant vectors were constructed based on the two longest open reading frames, which encoded 133 aa and 104 aa. A single nucleotide mutation introduced a premature stop codon in LAIR-MU1. A single nucleotide was deleted after the start codon (ATG) in LAIR-MU2. d, e Phenotypes of WT, LAIR, and transgenic plants carrying the LAIR mutant vectors. Growth traits of T3 transgenic lines were investigated, including mature plant phenotype (e) and primary panicle size (d). f Growth traits of LAIR-MU1 and LAIR-MU2 transgenic lines. Student’s t-test: *P < 0.05. Data are presented as the mean ± standard deviation (n = 12)