The function of lanthanides for methanotrophic and methylotrophic bacteria is gaining increasing attention, while knowledge about the role of rare earth elements (REEs) in nonmethylotrophic bacteria is still limited. The present study investigates the recently described differential expression of the two PQQ-EDHs of P. putida in response to lanthanides. We demonstrate that a specific TCS is crucial for their inverse regulation and provide evidence for a dual regulatory function of the LuxR-type response regulator involved. Thus, our study represents the first detailed characterization of the molecular mechanism underlying the REE switch of PQQ-EDHs in a nonmethylotrophic bacterium and stimulates subsequent investigations for the identification of additional genes or phenotypic traits that might be coregulated during REE-dependent niche adaptation.
KEYWORDS: lanthanides, LuxR-type regulator, PQQ, PedR2, PedS2, Pseudomonas putida, dehydrogenases, histidine kinase, periplasm, rare earth element switch, signal transduction, two-component regulatory systems
ABSTRACT
In Pseudomonas putida KT2440, two pyrroloquinoline quinone-dependent ethanol dehydrogenases (PQQ-EDHs) are responsible for the periplasmic oxidation of a broad variety of volatile organic compounds (VOCs). Depending on the availability of rare earth elements (REEs) of the lanthanide series (Ln3+), we have recently reported that the transcription of the genes encoding the Ca2+-utilizing enzyme PedE and the Ln3+-utilizing enzyme PedH are inversely regulated. With adaptive evolution experiments, site-specific mutations, transcriptional reporter fusions, and complementation approaches, we now demonstrate that the PedS2/PedR2 (PP_2671/PP_2672) two-component system (TCS) plays a central role in the observed REE-mediated switch of PQQ-EDHs in P. putida. We provide evidence that in the absence of lanthanum (La3+), the sensor histidine kinase PedS2 phosphorylates its cognate LuxR-type response regulator PedR2, which in turn not only activates pedE gene transcription but is also involved in repression of pedH. Our data further suggest that the presence of La3+ lowers kinase activity of PedS2, either by the direct binding of the metal ions to the periplasmic region of PedS2 or by an uncharacterized indirect interaction, leading to reduced levels of phosphorylated PedR2. Consequently, the decreasing pedE expression and concomitant alleviation of pedH repression causes—in conjunction with the transcriptional activation of the pedH gene by a yet unknown regulatory module—the Ln3+-dependent transition from PedE- to PedH-catalyzed oxidation of alcoholic VOCs.
IMPORTANCE The function of lanthanides for methanotrophic and methylotrophic bacteria is gaining increasing attention, while knowledge about the role of rare earth elements (REEs) in nonmethylotrophic bacteria is still limited. The present study investigates the recently described differential expression of the two PQQ-EDHs of P. putida in response to lanthanides. We demonstrate that a specific TCS is crucial for their inverse regulation and provide evidence for a dual regulatory function of the LuxR-type response regulator involved. Thus, our study represents the first detailed characterization of the molecular mechanism underlying the REE switch of PQQ-EDHs in a nonmethylotrophic bacterium and stimulates subsequent investigations for the identification of additional genes or phenotypic traits that might be coregulated during REE-dependent niche adaptation.
INTRODUCTION
In its natural soil habitat, Pseudomonas putida KT2440 is exposed to a broad range of potential carbon and energy sources (1–3), including plant-, fungus-, or bacterium-derived volatile organic compounds (VOCs) with alcohols or aldehydes as functional groups (4–6). For efficient capture and metabolism of such VOCs, P. putida makes use of two pyrroloquinoline quinone (PQQ)-dependent ethanol dehydrogenases (PQQ-EDHs)—namely, PedE and PedH—to carry out the initial oxidation steps in the periplasm of the cell (7, 8). In a recent study, we found that these two type I quinoproteins (9, 10) exhibit a similar substrate scope but require different metal cofactors (8). In contrast to PedE, which uses Ca2+ ions, PedH was characterized as a rare earth element (REE)-dependent enzyme that relies on the presence of lanthanides (Ln3+) for catalytic activity. Notably, due to their low solubility in most natural environments, REEs have long been considered to have no biological function (11). However, the discovery of the widespread occurrence of the REE-dependent XoxF type of PQQ-dependent methanol dehydrogenases (PQQ-MDHs), together with the more recent description of Ln3+-dependent PQQ-EDHs, has highlighted the important role of REEs for many bacterial species in various environmental compartments (8, 12–20).
While in the absence of Ln3+, the oxidation of methanol in methylotrophs is driven by Ca2+-dependent PQQ-MDHs (MxaF type), the presence of small amounts of REE ions is usually sufficient to trigger a transcriptional switch to the XoxF type of PQQ-MDHs. This inverse regulation, called the REE switch, has been reported for many methanotrophic and/or methylotrophic organisms (13, 16, 21–24). From the growing number of studies, it has become apparent that the molecular mechanism underlying this switch for PQQ-MDHs is complex and can substantially differ among species. For example, the inverse regulation in the nonmethanotrophic methylotroph Methylobacterium extorquens strain AM1 is controlled by two different two-component systems (TCSs) (MxcQE and MxbDM) and the orphan response regulator MxaB (25–27). In this organism, it has been found that the transcriptional activation of both enzymes, the Ca2+-dependent MxaF and the two Ln3+-dependent XoxF1 and XoxF2, is entirely lost in a ΔxoxF1 ΔxoxF2 double mutant (28). As a consequence, a complex regulation in which the different binding affinities of the apo form (no Ln3+ bound to the enzyme) and holo form (Ln3+ bound to the enzyme) of the XoxF proteins to the periplasmic domain of the sensor histidine kinase MxcQ is essential to regulate the switch was postulated (23).
The type I methanotroph Methylomicrobium buryatense strain 5GB1C is lacking homologues of the aforementioned TCS systems MxcQE and MxbDM (13). In this organism, the REE switch is regulated predominantly by the sensor histidine kinase MxaY (29). Chu and coworkers (29) found that the activation of MxaY in the absence of lanthanum activates transcription of the Ca2+-dependent enzyme MxaF by a so-far-unknown response regulator. In addition, the deletion of MxaY was found to almost entirely eliminate the Ln3+-mediated transcriptional activation of the Ln3+-dependent enzyme XoxF in a partially MxaB-dependent manner that also results in a severe growth defect, both in the presence and absence of Ln3+. As an additional layer of complexity, recent studies found that the presence of other metal ions such as copper, which is needed as a cofactor for the membrane-bound or particulate methane monooxygenase (pMMO) in methanotrophs, can significantly impact the REE-mediated switch (21, 22, 30).
In contrast to the increasing knowledge about the regulation of PQQ-MDHs in methylotrophs, the molecular basis underlying such a regulatory switch for PQQ-EDHs in nonmethylotrophic organisms is not established. In the present study, we identify the TCS encoded by PP_2671/PP_2672 (hereinafter referred to as PedS2/PedR2 according to the genetic nomenclature from Arias et al. [31]), consisting of the sensor histidine kinase PedS2 and its cognate LuxR-type transcriptional response regulator PedR2, as an essential regulatory module for the REE-mediated switch of PQQ-EDHs in P. putida KT2440. We provide evidence that the activity of PedS2 in the absence of lanthanides leads to phosphorylation of PedR2 (PedR2P), which serves a dual regulatory function. On the one hand, PedR2P acts as a strong transcriptional activator of the pedE gene, which is essential to allow growth with 2-phenylethanol in the absence of Ln3+. At the same time, PedR2P also functions as a repressor of pedH. From our data, we conclude that the presence of Ln3+ ions triggers a reduction in PedS2 activity, either by a direct binding of the metal to the periplasmic region of PedS2 or by an uncharacterized indirect interaction. This reduction of PedS2 activity, together with a proposed phosphatase activity of PedS2 under this condition, causes the accumulation of nonphosphorylated PedR2, which over time results in the loss of the regulatory activity of the protein, and facilitates—in concert with the positive-feedback function of PedH in the presence of Ln3+ ions—the switch between PedE- and PedH-dependent growth.
RESULTS
Identification of the sensor histidine kinase PedS2 as a lanthanide-responsive sensor.
As a consequence of the inverse regulation of pedE and pedH, a pedH deletion strain does not grow within 48 h with 2-phenylethanol as the sole carbon source in the presence of a critical concentration of La3+ in the culture medium (8). To test whether strains can evolve to overcome the repression of pedE in the presence of Ln3+, an adaptive evolution experiment was performed (see Fig. 1 for a general scheme).
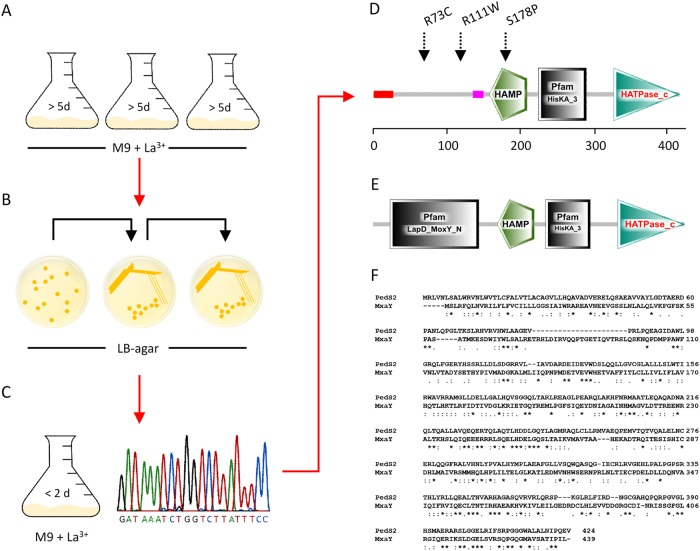
FIG 1 .
(A to D) Schemes of selection (A), clonal isolation (B), characterization and single nucleotide polymorphism (SNP) identification (C and D) in the two-component sensor histidine kinase PedS2 of the ΔpedHS1 (R73C), ΔpedHS2 (R111W), ΔpedHS3 (S178P) spontaneous mutants. (A) Cells of the ΔpedH strain were incubated in M9 medium supplemented with 5 mM 2-phenylethanol and 10 µM LaCl3 in plastic Erlenmeyer flasks (n = 3) at 30°C with shaking at 180 rpm. (B) After growth was observed (>5 days), dilutions from each culture were plated onto LB agar plates and incubated at 30°C. Individual clones were further streaked on LB agar twice prior to further characterization. (C) Clones were characterized for their growth behavior in M9 medium with 5 mM 2-phenylethanol in the presence of 10 µM LaCl3. Subsequently, one clone from each culture exhibiting faster growth than the parental ΔpedH strain was used for PCR amplification of the pedS2 gene and multiple-sequence alignment analysis with the native sequence of the gene from the Pseudomonas Genome Database (52). (D and E) Visualization of domain composition of PedS2 of P. putida (D) and MxaY of Methylomicrobium buryatense 5GB1C (E) using the prediction from the Simple Modular Architecture Research Tool (53). (F) Amino acid sequence alignment of the PedS2 and MxaY proteins generated with Clustal Omega (50).
When ΔpedH cultures were incubated with 2-phenylethanol in the presence of 10 µM La3+ ions for longer than 5 days, growth was observed, indicating the occurrence of adapted strains (data not shown). When independent clones were isolated from such cultures and passaged several times on LB agar medium, their growth phenotype with 2-phenylethanol was much faster (<2 days) than that observed for their ΔpedH parental strain and very similar to the growth phenotype of the wild-type strain KT2440. Similar spontaneous suppressor mutants have been reported in methylotrophic organisms such as Methylobacterium extorquens strain AM1, Methylomicrobium buryatense strain 5GB1C, and Methylobacterium aquaticum strain 22A during growth with methanol (13, 16, 29). In M. buryatense, whole-genome sequencing revealed that the suppressor mutant strain was characterized by a mutation in the membrane-bound two-component sensor histidine kinase MxaY (29). In Pseudomonas putida KT2440, the gene PP_2671 (hereinafter referred to as pedS2), located in close genomic proximity to pedE (PP_2674), encodes a membrane-bound histidine kinase sharing 25% amino acid sequence identity with MxaY (Fig. 1D to F). To test the hypothesis that mutations in pedS2 are responsible for the emergence of suppressor phenotypes in the ΔpedH mutant strain during growth in the presence of La3+, the gene was sequenced in three individually isolated mutants (ΔpedHS1 to ΔpedHS3 mutant strains) (Fig. 1A to D). This analysis revealed that all strains contained a mutation in the pedS2 sequence leading to a nonsynonymous substitution. These mutations were either located within a predicted periplasmic domain of unknown function (R73C [ΔpedHS1 strain] and R111W [ΔpedHS2 strain]) or within the HAMP domain (S178P [ΔpedHS3 strain]), which is responsible for signal transduction from the periplasm into the cytoplasm of the cell (32, 33). To verify that the identified mutations in pedS2 are the primary cause of the suppressor phenotype, the S178P mutation from the ΔpedHS3 strain was introduced into the genetic background of the ΔpedH strain, resulting in the ΔpedH_PedSS178P strain.
In subsequent growth experiments with 2-phenylethanol in the absence of La3+, the ΔpedH and ΔpedH_PedS2S178P mutants showed similar growth behavior with a lag phase of <32 h (Fig. 2A1 and A2). In the presence of 10 µM La3+, however, the ΔpedH strain showed no growth within 72 h, whereas the ΔpedH_PedS2S178P strain reached its maximum optical densities again after about 32 h of incubation, verifying that the observed mutation in the histidine kinase pedS2 gene was sufficient to cause the suppressor phenotype. We speculated that the LuxR-type response regulator exaE (PP_2672; hereinafter referred to as pedR2), which is located adjacent to pedS2 within the genome of P. putida KT2440, represents the target of PedS2 activity. This assumption is based on the fact that PedR2 represents a homologue (65% amino acid sequence identity) of EraR (ExaE; PA1980), which forms a two-component system (TCS) with the cytosolic histidine kinase EraS (ExaD; PA1979) that activates expression of the pedE homologue exaA in Pseudomonas aeruginosa (34, 35). To test this hypothesis, pedS2 as well as its potential cognate response regulator-encoding gene pedR2, were deleted in a ΔpedH background. In addition, strains suitable for probing promoter activity of pedE in ΔpedH, ΔpedH_PedS2S178P, ΔpedH ΔpedS2, and ΔpedH ΔpedR2 mutant strains were constructed and subsequently analyzed during growth with 2-phenylethanol in the presence and absence of La3+ (Fig. 2B).
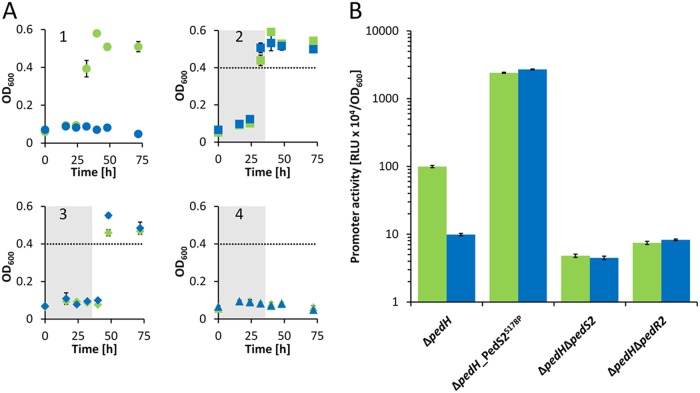
FIG 2 .
(A) Growth of ΔpedH, ΔpedH_pedS2S178P, ΔpedH ΔpedS2, and ΔpedH ΔpedR2 strains. The ΔpedH (circles; panel 1), ΔpedH_pedS2S178P (squares; panel 2), ΔpedH ΔpedS2 (diamonds; panel 3), and ΔpedH ΔpedR2 (triangles; panel 4) strains were grown at 30°C and 350 rpm shaking with M9 medium in 96-well plates supplemented with 5 mM 2-phenylethanol in the presence of 10 µM La3+ (blue symbols) or in the absence of La3+ (green symbols). The gray areas in panels 2 to 4 show the time point by which the parental ΔpedH strain (circles) reached an OD600 of >0.4 (dotted line). (B) Activities of the pedE promoter in ΔpedH, ΔpedH_PedS2S178P, ΔpedH ΔpedS2, and ΔpedH ΔpedR2 strains in the presence (blue bars) of 1 µM La3+ or absence of La3+ (green bars) or measured in M9 medium supplemented with 1 mM 2-phenylethanol. Promoter activities are presented in relative light units (RLU × 104) normalized to OD600. All data represent the means for biological triplicates, and error bars correspond to the respective standard deviations.
The PedS2/PedR2 TCS regulates pedE transcription in response to lanthanide availability.
In accordance with the observed growth patterns, pedE promoter activities in the ΔpedH_PedS2S178P mutant were almost identical in both the presence and absence of La3+ (ratio of pedE promoter activity in cells grown without La3+ to promoter activity in cells grown with 1 µM La3+, 0.89 ± 0.02) but increased more than 24-fold (24-fold ± 1-fold and 27-fold ± 1-fold, respectively) compared to the pedE promoter activities determined for cells of the ΔpedH strain grown in the absence of La3+ (Fig. 2B). The ΔpedH ΔpedS2 double mutant also showed a La3+-independent growth phenotype similar to that of the ΔpedH_PedS2S178P strain but with a clear delay (<48 h versus <32 h) (Fig. 2A3). Promoter activities for pedE were almost identical in this strain in the presence and absence of La3+ (ratio of pedE promoter activity in cells grown without La3+ to promoter activity in cells grown with 1 µM La3+, 1.07 ± 0.09) (Fig. 2B).
In contrast, incubations of 72 h (Fig. 2A4) or even prolonged incubations for more than 7 days (data not shown) did not result in detectable growth of the ΔpedH ΔpedR2 double mutant with 2-phenylethanol, both in the presence and absence of La3+. Correspondingly, pedE promoter activities in this strain were low compared to those observed for cells of the ΔpedH strain in the presence of La3+ but in a range similar to those observed for the ΔpedH ΔpedS2 strain in the presence and absence of La3+. These data demonstrate that the PedS2/PedR2 system is the predominant element in the La3+-dependent regulation of pedE and that PedR2 is essential for PedE-dependent growth. However, as PedE-dependent growth can still be observed in the absence of PedS2 after prolonged incubations and in a PedR2-dependent manner (Fig. 2A3 and A4), we assume that at least one additional lanthanide-independent kinase must be able to phosphorylate PedR2, leading to transcriptional activation of pedE and functional production of the calcium-dependent enzyme under these conditions.
The PedS2/PedR2 TCS regulates the partial repression of pedH in the absence of lanthanides.
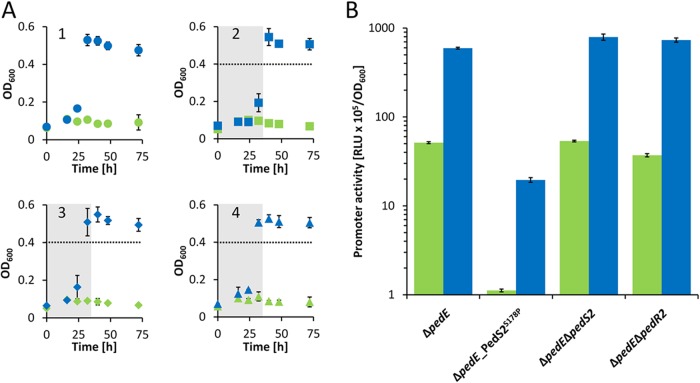
On the basis of the critical role of PedS2/PedR2 in the regulation of pedE and the fact that LuxR-type regulators have been demonstrated to be capable of acting as both transcriptional activators and repressors (36, 37), we speculated that this TCS could also be involved in the regulation of pedH. To test this hypothesis, ΔpedE_PedS2S178P, ΔpedE ΔpedS2, and ΔpedE ΔpedR2 mutant strains were generated, and a transcriptional reporter suitable for probing pedH promoter activities was integrated into the genome of each of these strains.
Experiments with 2-phenylethanol revealed that ΔpedE, ΔpedE ΔpedS2, and ΔpedE ΔpedR2 mutant strains showed La3+-dependent growth after a lag phase of <24 h and reached the maximum optical density at 600 nm (OD600) after ≤32 h (Fig. 3A1, A3, and A4). In contrast, the ΔpedE_PedS2S178P strain exhibited an extended lag phase (>24 h) and consistently reached the maximum OD600 only after prolonged incubations (≥32 h [Fig. 3A2]). In accordance with these growth results, the pedH promoter activities of ΔpedE, ΔpedE ΔpedS2, and ΔpedE ΔpedR2 strains were in a similar range, whereas pedH promoter activities in the ΔpedE_PedS2S178P strain were 45-fold ± 2-fold and 30-fold ± 2-fold lower than the promoter activities of the ΔpedE strain in the absence or presence of La3+, respectively (Fig. 3B). Assuming that the S178P mutation in PedS2 results in a sensor kinase activity that mimics that of the wild-type protein in the absence of lanthanides, it is very likely that PedS2 is responsible for the repression of pedH under the La3+-free conditions leading to the observed delay in growth. To find out whether this regulatory effect on pedH proceeds via the response regulator PedR2, as is the case for pedE, or is caused by an unknown additional target of PedS2, the ΔpedE_PedS2S178P ΔpedR2 triple mutant strain was generated and characterized for its growth phenotype (Fig. 4A). In this experiment, the ΔpedE and ΔpedE_PedS2S178P strains both grew with 2-phenylethanol but with clear differences in the corresponding lag phases and maximum growth rates (0.087 ± 0.003 h−1 versus 0.057 ± 0.001 h−1), confirming the previous results from growth in 96-well plates (Fig. 3A). In contrast, the additional deletion of the response regulator PedR2 eliminated the growth defect caused by the PedS2S178P allele, leading to a growth behavior of the ΔpedE_PedS2S178P ΔpedR2 strain (Fig. 4A), which was indistinguishable (maximum growth rate, 0.089 ± 0.003 h−1) from that of the ΔpedE strain.
FIG 3 .
(A) Growth of ΔpedE, ΔpedE_PedS2S178P, ΔpedE ΔpedS2, and ΔpedE ΔpedR2 strains. The ΔpedE (circles; panel 1), ΔpedE_PedS2S178P (squares; panel 2), ΔpedE ΔpedS2 (diamonds; panel 3), and ΔpedE ΔpedR2 (triangles; panel 4) were grown at 30°C and 350 rpm shaking with M9 medium in 96-well plates supplemented with 5 mM 2-phenylethanol in the presence of 10 µM La3+ (blue symbols) or absence of La3+ (green symbols). The gray areas in panels 2 to 4 show the time point by which the parental ΔpedE strain (circles) reached their maximum OD600. (B) Activities of the pedH promoter in ΔpedE, ΔpedE_PedS2S178P, ΔpedE ΔpedS2, and ΔpedE ΔpedR2 strains in the presence of 1 µM La3+ (blue bars) or in the absence of La3+ (green bars) or measured in M9 medium supplemented with 1 mM 2-phenylethanol. Promoter activities are presented in relative light units (RLU × 105) normalized to OD600. All data represent the means for biological triplicates, and error bars correspond to the respective standard deviations.
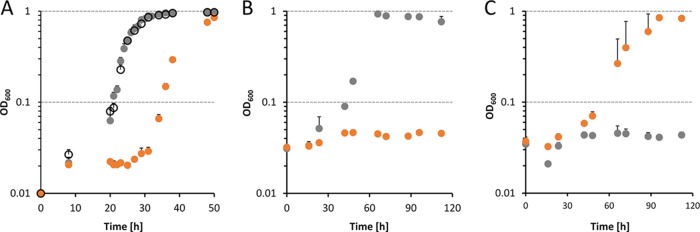
FIG 4 .
Growth of different P. putida strains at 30°C and 180 rpm shaking with M9 medium in polycarbonate Erlenmeyer flasks supplemented with 5 mM 2-phenylethanol and 10 µM La3+ in the absence (A) and presence of kanamycin (B and C) for plasmid maintenance. Flasks were inoculated at an OD600 of 0.01 (A) or 0.03 (B and C) with washed cells from M9 overnight cultures grown with succinate in the absence (A) or presence (B and C) of kanamycin and 0.2% (wt/vol) rhamnose to induce pJEM[PedR2] and pJEM[PedR2D53A] plasmids. (A) Growth of ΔpedE (black circles), ΔpedE_PedS2S178P (orange circles), and ΔpedE_PedS2S178P ΔpedR2 (gray circles) strains. (B) Growth of ΔpedH_PedS2S178P ΔpedR2 strain harboring pJEM[PedR2] (gray circles) or pJEM[PedR2D53A] (orange circles). (C) Growth of ΔpedE_PedS2S178P ΔpedR2 strain harboring pJEM[PedR2] (gray circles) or pJEM[PedR2D53A] (orange circles). Data points represent the means for biological triplets, and error bars correspond to the respective standard deviations (positive error values).
The conserved phosphorylation site D53 in PedR2 is essential for the REE-mediated switch.
In order to study the essentiality of the phosphorylation site at position D53 of PedR2 (38, 39), we used inducible low-copy-number constructs for the production of the wild-type PedR2 protein (pJEM[PedR2]) and a mutated variant, in which the conserved aspartate in the CheY-like receiver domain was replaced by an alanine (pJEM[PedR2D53A]). After transformation of these plasmids into the ΔpedH ΔpedR2 and ΔpedE_PedS2S178P ΔpedR2 mutant strains, their growth with 2-phenylethanol in the presence and absence of La3+ was monitored. When the plasmid-borne wild-type regulator PedR2 was induced in cells of the ΔpedH ΔpedR2 strain, growth was observed after a lag phase of <24 h (maximum growth rate, 0.032 ± 0.005 h−1), whereas the PedR2D53A variant was unable to restore PedE-dependent growth in the same strain (Fig. 4B). Intriguingly, the reverse result was obtained in the ΔpedE_PedS2S178P ΔpedR2 strain. Here, the PedR2D53A variant allowed PedH-dependent growth (maximum growth rate, 0.026 ± 0.002 h−1), whereas the wild-type regulator PedR2 did not lead to significant growth within 120 h of incubation (Fig. 4C).
DISCUSSION
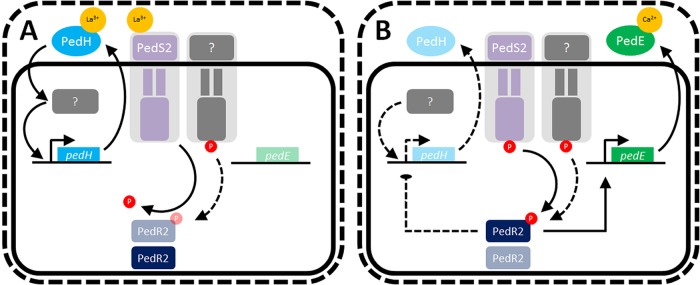
We recently demonstrated that in P. putida KT2440, the production of the two PQQ-EDHs PedE and PedH is both tightly and inversely regulated depending on lanthanide availability, representing the first reported REE switch for PQQ-EDHs in a nonmethylotrophic organism (8). In this study, we were able to show that Ln3+-dependent transcriptional activation of pedH is mostly, but not entirely, dependent on the presence of the PedH protein itself by a so-far unknown mechanism and transcriptional regulator (Fig. 5A). Notably, the Ln3+-dependent transcriptional repression of pedE remained elusive. In the current study, we present a detailed characterization of the mechanism underlying PedE and PedH regulation, in which the PedS2/PedR2 TCS acts as an essential signaling module for the REE-mediated switch between the two quinoproteins.
FIG 5 .
Working hypothesis of rare earth element (REE)-mediated switch of pedE and pedH in Pseudomonas putida KT2440. (A) The presence of REEs in the medium leads to binding of Ln3+ ions to PedH and the periplasmic domain of PedS2. The binding to PedH leads to the catalytic activation of the enzyme and triggers enhanced transcriptional activation of pedH by a so-far unknown mechanism and regulator, which is indicated by a gray box (8). Binding to the periplasmic domain of PedS2 leads to an outside-in signaling via the HAMP domain and decreases the kinase activity of the protein toward its cognate response regulator PedR2. In addition, this state of the PedS2 sensor is believed to exhibit phosphatase activity to reduce cross talk of an unidentified kinase (indicated by a gray membrane-bound protein) with activity toward PedR2 (for a more detailed explanation, see the text). (B) In the absence of REEs, the sensor kinase PedS2 is active and phosphorylates the cognate response regulator PedR2 at position D53. The phosphorylated PedR2 has a dual regulatory function as a strong pedE activator and a repressor of pedH. Black lines indicate known regulations or functionalities. Solid lines indicate a strong regulatory effect on genes or production of enzymes, whereas dotted lines indicate weaker regulatory effects or low production of enzymes.
Similar to the recently characterized spontaneous mutant of M. buryatense (29), we found that a single nonsynonymous mutation within the periplasmic region of the sensor histidine kinase PedS2 (PedS2S178P), which differs from the LapD/MoxY domain found in MxaY of M. buryatense (Fig. 1), is sufficient to terminate the Ln3+-mediated repression of pedE. Notably, various mutations at different sites of the protein can cause the observed suppressor phenotype. This might explain the repeated and fast occurrence of these suppressor mutants in our experiments and would support a similar notion in M. buryatense (13, 29).
Besides the essentiality for pedE regulation, our experimental data further provide strong evidence that PedS2 is also involved in the repression of pedH in the absence of lanthanides, but not in its Ln3+-dependent activation. This is based on the observation that the ΔpedE ΔpedS2 deletion strain did not show any differences in growth and pedH activation, while the ΔpedE_PedS2S178P suppressor mutant displayed decreased pedH promoter activity and a strongly increased lag phase in growth experiments in the presence of La3+. The complementation assay with the inducible PedR2 variants further demonstrates that the pedS2-dependent regulation of pedE and pedH is mediated by the LuxR-type response regulator PedR2 for both genes. From these data, we conclude that in the absence of La3+ ions, the PedS2 sensor histidine kinase is active and triggers phosphorylation of PedR2 at the conserved position D53 (PedR2P) (38, 39). The phosphorylated state of PedS2 subsequently has a dual regulatory function, namely, the activation of pedE and concomitant repression of pedH transcription (Fig. 5A). In this context, it is interesting to note that expression of the exaA gene in Azospirillum brasilense Sp7, which encodes a pyrroloquinoline quinone (PQQ)-dependent alcohol dehydrogenase, is dependent on σ54 and its interaction with a LuxR-type response regulator that shares 45% sequence identity with PedR2 (40). Whether the transcriptional activation of pedE is dependent on a similar interaction of PedR2 with a specific sigma factor is currently unknown.
To our surprise, a strain lacking PedS2 (ΔpedH ΔpedS2 strain) was still able to grow on 2-phenylethanol in the absence of Ln3+, even though it exhibited an increased lag phase and low pedE promoter activities (Fig. 2A3). As our study provides strong evidence that phosphorylation of PedR2 is essential for transcriptional activation of pedE, this observation indicates that an additional so-far unidentified kinase beside PedS2 is capable of phosphorylating PedR2 to facilitate growth of this strain under these conditions (indicated in Fig. 5 as a gray membrane-bound protein). Given that such an additional kinase exists, it is even more surprising that a pedH single mutant, in contrast to the aforementioned ΔpedH ΔpedS2 double mutant, is able to grow in the presence of REEs only when PedS2 is mutated (e.g., PedS2S178P [Fig. 2A1]). This suggests that the activity of the additional kinase toward PedR2 in the presence of La3+ is repressed as long as PedS2 is functional. As an intrinsic phosphatase activity has been found for many bacterial sensory histidine kinases (33, 41, 42), we hence propose that PedS2 also exhibits phosphatase activity on PedR2P in the presence of La3+, thereby ensuring specificity of the signal transduction pathway and eliminating interference from other nonspecific kinases. In our working hypothesis, the presence of lanthanides in the medium leads to the repression of PedS2 kinase activity, most likely by direct binding of the metal ions to its periplasmic domain. The reduced kinase activity and postulated phosphatase activity of PedS2 in the presence of Ln3+ consequently leads to the accumulation of unphosphorylated PedR2, which finally results in the loss of its regulatory functions. In addition, the transcription of pedH is activated via a yet unknown pathway, in which a functional PedH protein is an essential component, most likely by acting as a lanthanide sensor (8).
In conclusion, it appears that the REE-mediated switches in Methylobacterium extorquens AM1 and Methylomicrobium buryatense during growth with methanol are predominantly dependent on only one lanthanide-responsive pathway, which either proceeds via the XoxF1 and XoxF2 proteins or via the MxaY protein (23, 29). Our results establish that in P. putida KT2440, a combination of at least two independent pathways are important to orchestrate the inverse regulation of pedE and pedH in response to lanthanides efficiently.
Several recent studies suggest that in methano- and methylotrophic bacteria, the REE switch might affect more genes than only the genes needed for the periplasmic oxidation system itself (16, 22, 43). Reports on physiological consequences are, however, inconsistent as some studies found no effects (13, 16, 23, 44), whereas other studies reported a stimulating effect on biofilm formation, growth rates, and overall yields in the presence of REE (43, 45). We think it is not unlikely that additional REE-mediated regulatory effects also exist in P. putida KT2440 in a context-dependent manner. Thus, one of our current foci is to investigate the global regulatory impact and physiological consequences of the presence and absence of lanthanides under various environmental conditions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
A detailed description of the bacterial strains, plasmids, and primers used in this study can be found in Tables 1 and 2. If not stated otherwise, Escherichia coli and Pseudomonas putida KT2440 strains were maintained on solidified LB medium. Routinely, strains were cultured in liquid LB medium (46) or a modified M9 salt medium (8) supplemented with 25 mM succinate or 5 mM 2-phenylethanol as a source of carbon and energy at 30°C and shaking, if not stated otherwise. For maintenance and selection, 40 µg ml−1 kanamycin or 15 µg ml−1 gentamicin for E. coli or 40 µg ml−1 kanamycin, 20 µg ml−1 5-fluorouracil, or 15 µg ml−1 gentamicin for P. putida strains was added to the medium, if indicated.
TABLE 1 .
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant feature(s) | Reference |
|---|---|---|
| Bacterial strains | ||
| KT2440 | Wild-type strain of Pseudomonas putida (ATCC 47054) | |
| KT2440* | KT2440 with a markerless deletion of upp; parent strain for deletion mutants | 48 |
| ΔpedE | KT2440* with a markerless deletion of pedE | 7 |
| ΔpedE ΔpedS2 | ΔpedE strain with a markerless deletion of pedS2 (PP_2671) | This study |
| ΔpedE ΔpedR2 | ΔpedE strain with a markerless deletion of pedR2 (PP_2672) | This study |
| ΔpedE_PedS2S178P | ΔpedE strain with S178P mutation in PedS2 | This study |
| ΔpedE_PedS2S178P ΔpedR2 | ΔpedE_PedS2S178P strain with a markerless deletion of pedR2 | This study |
| ΔpedH | KT2440* with a markerless deletion of pedH | 7 |
| ΔpedH ΔpedS2 | ΔpedH strain with a markerless deletion of pedS2 | This study |
| ΔpedH ΔpedR2 | ΔpedH strain with a markerless deletion of pedR2 | This study |
| ΔpedH_PedS2S178P | ΔpedH strain with S178P mutation PedS2 | This study |
| ΔpedH_PedS2S178P ΔpedR2 | ΔpedH_PedS2S178P strain with a markerless deletion of pedR2 | This study |
| E. coli BL21(DE3) | F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | |
| E. coli DH5α | fhuA2 lac(del)U169 phoA glnV44 φ80′ lacZ(del)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | |
| KT2440*::Tn7-pedE-lux | KT2440* with insertion of miniTn7-pedE-lux | 8 |
| KT2440*::Tn7-pedH-lux | KT2440* with insertion of miniTn7-pedH-lux | 8 |
| ΔpedE::Tn7-pedE-lux | ΔpedE strain with insertion of miniTn7-pedE-lux | 8 |
| ΔpedE::Tn7-pedH-lux | ΔpedE strain with insertion of miniTn7-pedH-lux | 8 |
| ΔpedH::Tn7-pedE-lux | ΔpedH strain with insertion of miniTn7-pedE-lux | 8 |
| ΔpedH::Tn7-pedH-lux | ΔpedH strain with insertion of miniTn7-pedH-lux | 8 |
| ΔpedE ΔpedS2::Tn7-pedH-lux | ΔpedE ΔpedS2 strain with insertion of miniTn7-pedH-lux | This study |
| ΔpedE ΔpedR2::Tn7-pedH-lux | ΔpedE ΔpedR2 strain with insertion of miniTn7-pedH-lux | This study |
| ΔpedE_PedS2S178P::Tn7-pedH-lux | ΔpedE_PedS2S178P strain with insertion of miniTn7-pedH-lux | This study |
| ΔpedH ΔpedS2::Tn7-pedE-lux | ΔpedH ΔpedS2 strain with insertion of miniTn7-pedE-lux | This study |
| ΔpedH ΔpedR2::Tn7-pedE-lux | ΔpedH ΔpedR2 strain with insertion of miniTn7-pedE-lux | This study |
| ΔpedH_PedS2S178P::Tn7-pedE-lux | ΔpedH_PedS2S178P strain with insertion of miniTn7-pedE-lux | This study |
| Plasmids | ||
| pJeM1 | Rhamnose-inducible vector (pBBR1MCS backbone) with low copy number | 51 |
| pJOE6261.2 | Suicide vector for gene deletions | 48 |
| pMW55 | pJOE6261.2-based deletion vector for gene PP_2671 | This study |
| pMW56 | pJOE6261.2-based vector for introducing the S178P mutation in PedS2 | This study |
| pMW61 | pJOE6261.2-based deletion vector for gene PP_2672 | This study |
| pUC18-mini-Tn7T-pedE-lux-Gm | pUC18-mini-Tn7T-Gm-lux vector with the promoter of pedE driving transcription of luxCDABE | 8 |
| pUC18-mini-Tn7T-pedH-lux-Gm | pUC18-mini-Tn7T-lux-Gm vector with the promoter of pedH driving transcription of luxCDABE | 8 |
| pTNS2 | Helper plasmid for Tn7 integration | 49 |
| pJEM[PedR2] | Rhamnose-inducible induction of PedR2 | This study |
| pJEM[PedR2D53A] | Rhamnose-inducible induction of PedR2D53A | This study |
TABLE 2 .
Primers used in this study
| Primer | Primer sequence (5′ → 3′) | Annealing temp (°C) |
|---|---|---|
| MWH85 | GGAAATATGCAGAAAGTAGCGCTCG | 60 |
| MWH86 | TCTTCACCACTGGCGGCCT | 60 |
| MWH90 | GCCGCTTTGGTCCCGCAGGCACTGGCTGCTGC | 60 |
| MWH91 | CGATATTCAAAGCGGTTCTCCTCAGGC | 60 |
| MWH92 | GAACCGCTTTGAATATCGTGTTGGTCGATGACCAC | 60 |
| MWH93 | GCAGGTCGACTCTAGAGGATGCACAAGCTCGGCG | 60 |
| MWH98 | GCCGCTTTGGTCCCGAGGTAGTAATTCAGTGCGGGGG | 60 |
| MWH99 | TGCCCGCCTGGGACCTGGTG | 60 |
| MWH100 | GGTCCCAGGCGGGCAATTG | 60 |
| MWH101 | GCAGGTCGACTCTAGAGGCAGCCATTGTCGCGAATG | 60 |
| MWH106 | GCCGCTTTGGTCCCGGCAGGAGCAGGAGCGTAC | 65 |
| MWH107 | TGAAATACCCACACCTCCTGGGGAATGTTAAG | 65 |
| MWH108 | GGAGGTGTGGGTATTTCATTGCACCTGTTGGGGC | 65 |
| MWH109 | GCAGGTCGACTCTAGAGGAGCCAACCTGACCC | 65 |
Liquid medium growth experiments.
All liquid growth experiments were carried out using modified M9 medium with 25 mM succinate or 5 mM 2-phenylethanol as the sole source of carbon and energy (see above) in 125-ml or 250-ml polycarbonate Erlenmeyer flasks (Corning) or in 96-well 2-ml deep-well plates (Carl Roth) as described previously (8). Briefly, washed cells from overnight cultures grown with succinate at 30°C and 180 rpm shaking were used to inoculate fresh medium with an optical density at 600 nm (OD600) of 0.01 and incubated at 30°C and 180 rpm (growth experiments in polycarbonate Erlenmeyer flasks) or 350 rpm (growth experiments in 96-well plates) shaking. Maximum growth rates were calculated from three time points during the exponential phase of growth.
Construction of plasmids.
For construction of the deletion plasmids pMW55, pMW56, and pMW61, the 600-bp regions upstream and downstream of the pedS2 gene (PP_2671) or amino acid residue S178 in the pedS2 gene (PP_2671) or pedR2 gene (PP_2672) were amplified from genomic DNA of P. putida KT2440 using primers MWH90 to MWH93, MWH98 to MWH101, or MWH106 to MWH109 (Table 2). The two up- and downstream fragments and BamHI-digested pJOE6261.2 were then joined together using one-step isothermal assembly (47). Upon subsequent transformation of the constructs into E. coli BL21(DE3) cells, the correctness of the plasmids was confirmed by Sanger sequencing. Plasmids pJEM[PedR2] and pJEM[PedR2D53A] encoding PedR2 or PedR2 with mutated amino acid residue 53 (D→A) under a rhamnose-inducible promoter were ordered from an external source (Eurofins).
Strain constructions and isolation of suppressor mutants.
The pedS2 (PP_2671) and pedR2 (PP_2672) negative mutants as well as the PedS2S178P allele were constructed using a recently described system for markerless gene deletion in P. putida KT2440 (48). Briefly, the integration vectors harboring the up- and downstream regions of the target genes (pMW55 and pMW61) or the up- and downstream regions of the region to be mutated, including the desired S178P mutation (pMW56) were transformed into P. putida KT2440* and kanamycin-resistant (Kanr) and 5-fluorouracil-sensitive (5-FUs) clones were selected on LB Kan agarose plates. After incubation at 30°C for 24 h in LB medium without selection markers, clones that were 5-FU resistant (5-FUr) and Kans were tested for successful gene deletion using primer pair MWH90/MWH93 or MWH106/MWH109 for the pedS2 or pedR2 gene, respectively. The presence of the underlying PedS2S178P mutation was verified by Sanger sequencing after the pedS2 gene of 5-FUr and Kans clones was amplified using primer pair MWH85/MWH86. ΔpedH suppressor mutant strains were isolated from 25-ml liquid M9 cultures with 5 mM 2-phenylethanol as the sole source of carbon and energy supplemented with 10 µM La3+ upon >5-day incubation at 30°C and 180 rpm. Strains were passaged three times in LB agar plates and reevaluated for growth in liquid M9 medium supplemented with 5 mM 2-phenylethanol and 10 µM La3+. From cultures that showed growth with a lag phase similar to that of the wild-type KT2440* strain, the pedS2 gene was amplified by PCR using primer pair M85/M86, and mutations were identified by Sanger sequencing.
To construct reporter strains for the analysis of pedE and pedH promoter activity in different genetic backgrounds, plasmids pUC18-mini-Tn7T-pedE-lux-Gm and pUC18-mini-Tn7T-pedH-lux-Gm (8) were coelectroporated with the helper plasmid pTNS2 into selected mutant strains of P. putida KT2440 (Table 1). Proper chromosomal integration of the mini-Tn7 element in gentamicin-resistant transformants was verified by colony PCR using Pput-glmSDN and PTn7R primers as described previously (49).
Reporter gene fusion assays.
For quantitative measurement of pedE and pedH promoter activity, strains of P. putida harboring a Tn7-based pedE-lux or pedH-lux transcriptional reporter fusion were grown overnight in LB medium with gentamicin (15 µg ⋅ ml−1), diluted to an OD600 of 0.2 in fresh LB medium, and grown to an OD600 of 0.6. The cells were then washed three times in M9 medium without a carbon source and finally adjusted to an OD600 of 0.2 in M9 medium with 1 mM 2-phenylethanol. For luminescence measurements, 198 µl of a cell suspension was added to 2 µl of a 100 µM LaCl3 solution in white 96-well plates with a clear bottom (µClear; Greiner Bio-One). Microtiter plates were placed in a humid box to prevent evaporation and incubated at 28°C with continuous agitation (180 rpm), and light emission and OD600 were recorded at regular intervals in an FLX-Xenius plate reader (SAFAS, Monaco) for 6 h. For both parameters, the background provided by the M9 medium was subtracted, and the luminescence was normalized to the corresponding OD600. Experiments were performed with biological triplicates, and data are presented as the mean values with error bars representing the corresponding standard deviations.
Sequence identity determination.
Protein sequence identities were determined based on amino acid sequence alignments of the proteins of interest generated using the Clustal Omega multiple-sequence alignment tool (50).
ACKNOWLEDGMENTS
The work of Matthias Wehrmann and Janosch Klebensberger was supported by an individual research grant from the Deutsche Forschungsgemeinschaft (DFG) (KL 2340/2-1). The work of Charlotte Berthelot and Patrick Billard was supported in part by Labex Resources 21 (ANR-10-LABX-21-01).
We thank Bernhard Hauer for his continuous support.
We declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
REFERENCES
- 1.Regenhardt D, Heuer H, Heim S, Fernandez DU, Strömpl C, Moore ERB, Timmis KN. 2002. Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ Microbiol 4:912–915. doi: 10.1046/j.1462-2920.2002.00368.x. [DOI] [PubMed] [Google Scholar]
- 2.Wackett LP. 2003. Pseudomonas putida —a versatile biocatalyst. Nat Biotechnol 21:136–138. doi: 10.1038/nbt0203-136. [DOI] [PubMed] [Google Scholar]
- 3.Wu X, Monchy S, Taghavi S, Zhu W, Ramos J, van der Lelie D. 2011. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol Rev 35:299–323. doi: 10.1111/j.1574-6976.2010.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Insam H, Seewald MSA. 2010. Volatile organic compounds (VOCs) in soils. Biol Fertil Soils 46:199–213. doi: 10.1007/s00374-010-0442-3. [DOI] [Google Scholar]
- 5.Peñuelas J, Asensio D, Tholl D, Wenke K, Rosenkranz M, Piechulla B, Schnitzler JP. 2014. Biogenic volatile emissions from the soil. Plant Cell Environ 37:1866–1891. doi: 10.1111/pce.12340. [DOI] [PubMed] [Google Scholar]
- 6.van Dam NM, Weinhold A, Garbeva P. 2016. Calling in the dark: the role of volatiles for communication in the rhizosphere, p 175–210. In Blande JD, Glinwood R (ed), Deciphering chemical language of plant communication. Springer International, Cham, Switzerland. [Google Scholar]
- 7.Mückschel B, Simon O, Klebensberger J, Graf N, Rosche B, Altenbuchner J, Pfannstiel J, Huber A, Hauer B. 2012. Ethylene glycol metabolism by Pseudomonas putida. Appl Environ Microbiol 78:8531–8539. doi: 10.1128/AEM.02062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wehrmann M, Billard P, Martin-Meriadec A, Zegeye A, Klebensberger J. 2017. Functional role of lanthanides in enzymatic activity and transcriptional regulation of pyrroloquinoline quinone-dependent alcohol dehydrogenases in Pseudomonas putida KT2440. mBio 8:e00570-17. doi: 10.1128/mBio.00570-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toyama H, Mathews FS, Adachi O, Matsushita K. 2004. Quinohemoprotein alcohol dehydrogenases: structure, function, and physiology. Arch Biochem Biophys 428:10–21. doi: 10.1016/j.abb.2004.03.037. [DOI] [PubMed] [Google Scholar]
- 10.Anthony C. 2001. Pyrroloquinoline quinone (PQQ) and quinoprotein enzymes. Antioxid Redox Signal 3:757–774. doi: 10.1089/15230860152664966. [DOI] [PubMed] [Google Scholar]
- 11.Sarkar B. 2002. Heavy metals in the environment. CRC Press, Boca Raton, FL. [Google Scholar]
- 12.Fitriyanto NA, Fushimi M, Matsunaga M, Pertiwiningrum A, Iwama T, Kawai K. 2011. Molecular structure and gene analysis of Ce3+-induced methanol dehydrogenase of Bradyrhizobium sp. MAFF211645. J Biosci Bioeng 111:613–617. doi: 10.1016/j.jbiosc.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Chu F, Lidstrom ME. 2016. XoxF acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense. J Bacteriol 198:1317–1325. doi: 10.1128/JB.00959-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Good NM, Vu HN, Suriano CJ, Subuyuj GA, Skovran E, Martinez-Gomez NC. 2016. Pyrroloquinoline quinone ethanol dehydrogenase in Methylobacterium extorquens AM1 extends lanthanide-dependent metabolism to multicarbon substrates. J Bacteriol 198:3109–3118. doi: 10.1128/JB.00478-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pol A, Barends TRM, Dietl A, Khadem AF, Eygensteyn J, Jetten MSM, Op den Camp HJM. 2014. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ Microbiol 16:255–264. doi: 10.1111/1462-2920.12249. [DOI] [PubMed] [Google Scholar]
- 16.Masuda S, Suzuki Y, Fujitani Y, Mitsui R, Nakagawa T, Shintani M, Tani A. 2018. Lanthanide-dependent regulation of methylotrophy in Methylobacterium aquaticum strain 22A. mSphere 3:e00462-17. doi: 10.1128/mSphere.00462-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vekeman B, Speth D, Wille J, Cremers G, De Vos P, Op den Camp HJM, Heylen K. 2016. Genome characteristics of two novel type I methanotrophs enriched from North Sea sediments containing exclusively a lanthanide-dependent XoxF5-type methanol dehydrogenase. Microb Ecol 72:503–509. doi: 10.1007/s00248-016-0808-7. [DOI] [PubMed] [Google Scholar]
- 18.Lv H, Sahin N, Tani A. 2018. Isolation and genomic characterization of Novimethylophilus kurashikiensis gen. nov. sp. nov., a new lanthanide-dependent methylotrophic species of Methylophilaceae. Environ Microbiol 20:1204–1223. doi: 10.1111/1462-2920.14062. [DOI] [PubMed] [Google Scholar]
- 19.Shiller AM, Chan EW, Joung DJ, Redmond MC, Kessler JD. 2017. Light rare earth element depletion during Deepwater Horizon blowout methanotrophy. Sci Rep 7:10389. doi: 10.1038/s41598-017-11060-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taubert M, Grob C, Howat AM, Burns OJ, Dixon JL, Chen Y, Murrell JC. 2015. XoxF encoding an alternative methanol dehydrogenase is widespread in coastal marine environments. Environ Microbiol 17:3937–3948. doi: 10.1111/1462-2920.12896. [DOI] [PubMed] [Google Scholar]
- 21.Farhan Ul Haque M, Kalidass B, Bandow N, Turpin EA, DiSpirito AA, Semrau JD. 2015. Cerium regulates expression of alternative methanol dehydrogenases in Methylosinus trichosporium OB3b. Appl Environ Microbiol 81:7546–7552. doi: 10.1128/AEM.02542-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu W, Semrau JD. 2017. Copper and cerium-regulated gene expression in Methylosinus trichosporium OB3b. Appl Microbiol Biotechnol 101:8499–8516. doi: 10.1007/s00253-017-8572-2. [DOI] [PubMed] [Google Scholar]
- 23.Vu HN, Subuyuj GA, Vijayakumar S, Good NM, Martinez-Gomez NC, Skovran E. 2016. Lanthanide-dependent regulation of methanol oxidation systems in Methylobacterium extorquens AM1 and their contribution to methanol growth. J Bacteriol 198:1250–1259. doi: 10.1128/JB.00937-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Y, Huang J, Zhao F, Chistoserdova L. 2018. Physiological effect of XoxG(4) on lanthanide-dependent methanotrophy. mBio 9:e02430-17. doi: 10.1128/mBio.02430-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Springer AL, Auman AJ, Lidstrom ME. 1998. Sequence and characterization of mxaB, a response regulator involved in regulation of methanol oxidation, and of mxaW, a methanol-regulated gene in Methylobacterium extorquens AM1. FEMS Microbiol Lett 160:119–124. doi: 10.1111/j.1574-6968.1998.tb12900.x. [DOI] [PubMed] [Google Scholar]
- 26.Springer AL, Morris CJ, Lidstrom ME. 1997. Molecular analysis of mxbD and mxbM, a putative sensor-regulator pair required for oxidation of methanol in Methylobacterium extorquens AM1. Microbiology 143:1737–1744. doi: 10.1099/00221287-143-5-1737. [DOI] [PubMed] [Google Scholar]
- 27.Xu HH, Janka JJ, Viebahn M, Hanson RS. 1995. Nucleotide sequence of the mxcQ and mxcE genes, required for methanol dehydrogenase synthesis in Methylobacterium organophilum XX: a two-component regulatory system. Microbiology 141:2543–2551. doi: 10.1099/13500872-141-10-2543. [DOI] [PubMed] [Google Scholar]
- 28.Skovran E, Palmer AD, Rountree AM, Good NM, Lidstrom ME. 2011. XoxF is required for expression of methanol dehydrogenase in Methylobacterium extorquens AM1. J Bacteriol 193:6032–6038. doi: 10.1128/JB.05367-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu F, Beck DAC, Lidstrom ME. 2016. MxaY regulates the lanthanide-mediated methanol dehydrogenase switch in Methylomicrobium buryatense. PeerJ 4:e2435. doi: 10.7717/peerj.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Semrau JD, DiSpirito AA, Gu W, Yoon S. 2018. Metals and methanotrophy. Appl Environ Microbiol 84:e02289-17. doi: 10.1128/AEM.02289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arias S, Olivera ER, Arcos M, Naharro G, Luengo JM. 2008. Genetic analyses and molecular characterization of the pathways involved in the conversion of 2-phenylethylamine and 2-phenylethanol into phenylacetic acid in Pseudomonas putida U. Environ Microbiol 10:413–432. doi: 10.1111/j.1462-2920.2007.01464.x. [DOI] [PubMed] [Google Scholar]
- 32.Parkinson JS. 2010. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu Rev Microbiol 64:101–122. doi: 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- 33.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 34.Schobert M, Görisch H. 2001. A soluble two-component regulatory system controls expression of quinoprotein ethanol dehydrogenase (QEDH) but not expression of cytochrome c(550) of the ethanol-oxidation system in Pseudomonas aeruginosa. Microbiology 147:363–372. doi: 10.1099/00221287-147-2-363. [DOI] [PubMed] [Google Scholar]
- 35.Mern DS, Ha SW, Khodaverdi V, Gliese N, Görisch H. 2010. A complex regulatory network controls aerobic ethanol oxidation in Pseudomonas aeruginosa: indication of four levels of sensor kinases and response regulators. Microbiology 156:1505–1516. doi: 10.1099/mic.0.032847-0. [DOI] [PubMed] [Google Scholar]
- 36.Van Kessel JC, Rutherford ST, Shao Y, Utria AF, Bassler BL. 2013. Individual and combined roles of the master regulators AphA and LuxR in control of the Vibrio harveyi quorum-sensing regulon. J Bacteriol 195:436–443. doi: 10.1128/JB.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waters CM, Bassler BL. 2006. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev 20:2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milani M, Leoni L, Rampioni G, Zennaro E, Ascenzi P, Bolognesi M. 2005. An active-like structure in the unphosphorylated StyR response regulator suggests a phosphorylation-dependent allosteric activation mechanism. Structure 13:1289–1297. doi: 10.1016/j.str.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 39.Bourret RB, Hess JF, Simon MI. 1990. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci U S A 87:41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh VS, Dubey AP, Gupta A, Singh S, Singh BN, Tripathi AK. 2017. Regulation of a glycerol-induced quinoprotein alcohol dehydrogenase by σ54 and a LuxR-type regulator in Azospirillum brasilense Sp7. J Bacteriol 199:e00035-17. doi: 10.1128/JB.00035-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowland MA, Deeds EJ. 2014. Crosstalk and the evolution of specificity in two-component signaling. Proc Natl Acad Sci U S A 111:5550–5555. doi: 10.1073/pnas.1317178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agrawal R, Sahoo BK, Saini DK. 2016. Cross-talk and specificity in two-component signal transduction pathways. Future Microbiol 11:685–697. doi: 10.2217/fmb-2016-0001. [DOI] [PubMed] [Google Scholar]
- 43.Good NM, Walser ON, Moore RS, Suriano C, Huff AF, Martinez-Gomez NC. 2018. Investigation of lanthanide-dependent methylotrophy uncovers complementary roles for alcohol dehydrogenase enzymes. bioRxiv 10.1101/329011. [DOI]
- 44.Nakagawa T, Mitsui R, Tani A, Sasa K, Tashiro S, Iwama T, Hayakawa T, Kawai K. 2012. A catalytic role of XoxF1 as La3+-dependent methanol dehydrogenase in Methylobacterium extorquens strain AM1. PLoS One 7:e50480. doi: 10.1371/journal.pone.0050480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fitriyanto NA, Nakamura M, Muto S, Kato K, Yabe T, Iwama T, Kawai K, Pertiwiningrum A. 2011. Ce3+-induced exopolysaccharide production by Bradyrhizobium sp. MAFF211645. J Biosci Bioeng 111:146–152. doi: 10.1016/j.jbiosc.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 46.Maniatis T, Sambrook J, Fritsch EF. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 47.Gibson DG. 2011. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graf N, Altenbuchner J. 2011. Development of a method for markerless gene deletion in Pseudomonas putida. Appl Environ Microbiol 77:5549–5552. doi: 10.1128/AEM.05055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 50.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeske M, Altenbuchner J. 2010. The Escherichia coli rhamnose promoter rhaP BAD is in Pseudomonas putida KT2440 independent of Crp–cAMP activation. Appl Microbiol Biotechnol 85:1923–1933. doi: 10.1007/s00253-009-2245-8. [DOI] [PubMed] [Google Scholar]
- 52.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FSL. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Letunic I, Bork P. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]