FIG 1 .
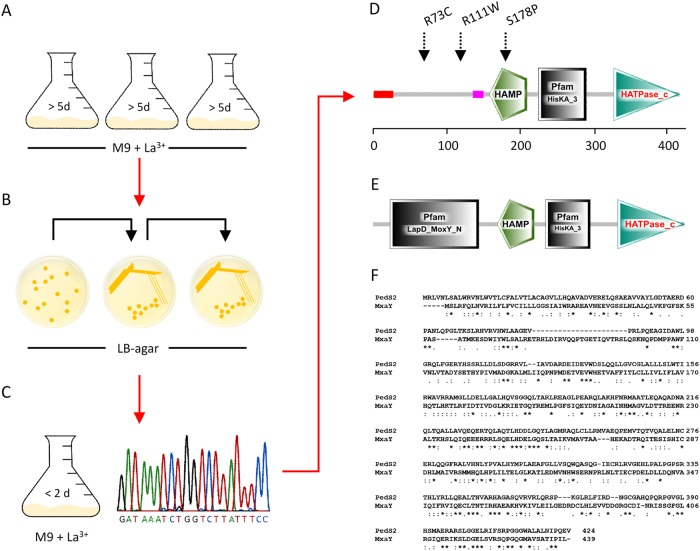
(A to D) Schemes of selection (A), clonal isolation (B), characterization and single nucleotide polymorphism (SNP) identification (C and D) in the two-component sensor histidine kinase PedS2 of the ΔpedHS1 (R73C), ΔpedHS2 (R111W), ΔpedHS3 (S178P) spontaneous mutants. (A) Cells of the ΔpedH strain were incubated in M9 medium supplemented with 5 mM 2-phenylethanol and 10 µM LaCl3 in plastic Erlenmeyer flasks (n = 3) at 30°C with shaking at 180 rpm. (B) After growth was observed (>5 days), dilutions from each culture were plated onto LB agar plates and incubated at 30°C. Individual clones were further streaked on LB agar twice prior to further characterization. (C) Clones were characterized for their growth behavior in M9 medium with 5 mM 2-phenylethanol in the presence of 10 µM LaCl3. Subsequently, one clone from each culture exhibiting faster growth than the parental ΔpedH strain was used for PCR amplification of the pedS2 gene and multiple-sequence alignment analysis with the native sequence of the gene from the Pseudomonas Genome Database (52). (D and E) Visualization of domain composition of PedS2 of P. putida (D) and MxaY of Methylomicrobium buryatense 5GB1C (E) using the prediction from the Simple Modular Architecture Research Tool (53). (F) Amino acid sequence alignment of the PedS2 and MxaY proteins generated with Clustal Omega (50).