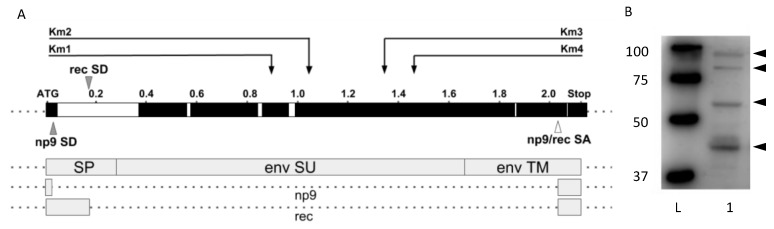
Figure 1.
Schematic representation of the human endogenous retrovirus (HERV)-Kenv gene and of the spliced mRNAs, showing the targets for gRNAs, and the basal production of HERV-K Env proteins by the LNCaP cell line. (A) Map of the proviral HERV-Kenv gene, spanning from the ATG to the Stop codon. The singly spliced SU and TM env mRNA, the doubly spliced np9 and rec mRNAs are shown as white boxes. The arrows indicate the targets of the Km1, Km2, Km3, and Km4 gRNAs. The arrowheads indicate the donor (grey arrowhead) and acceptor (white arrowhead) splicing sites, leading to the HERV-Knp9 and HERV-Krec mRNA splicing. (B) HERV-K Env proteins produced by LNCaP cells cultured for 48 h, as detected by Western blot assay. After blotting, the relevant bands were recognized by an anti-HERV-K Env mAb, as described in Methods. See Results for details. Lane 1: protein ladder (precision plus protein western C standards, Biorad, Hercules, California, USA), Lane 2: LNCaP cell lysate. Arrowheads: Env-specific bands. All the four HERV-K Env species are visible in this membrane.