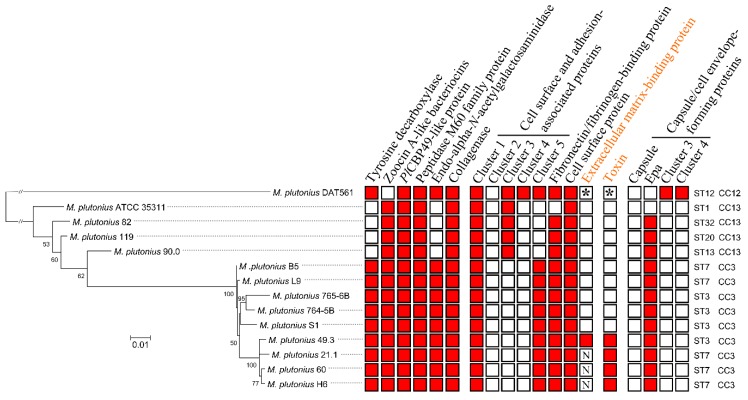
Figure 4.
Gene content tree based on presence or absence of orthologous proteins. For constructing the phylogenetic tree, a presence/absence binary matrix was created from orthologous groups to calculate phylogeny with RAxML v8.1.3 [31]. The atypical strain M. plutonius DAT561 was used as the outgroup. Numbers at nodes are bootstrap values calculated from 1000 re-samplings to generate a majority consensus tree. The scale bar indicates divergence in presence or absence of proteins. STs are shown on the right. Color-filled boxes to the right of the organisms show the presence of the indicated proteins. Genes encoding putative virulence factors in red font are located on pMP19. An ‘N’ symbolizes that the respective open reading frame (ORF) is not complete due to a gap in the DNA sequence. An asterisk symbolizes that the respective ORF has been identified only in the second version of the DAT561 genome [47]. (Epa: enterococcal polysaccharide antigen).