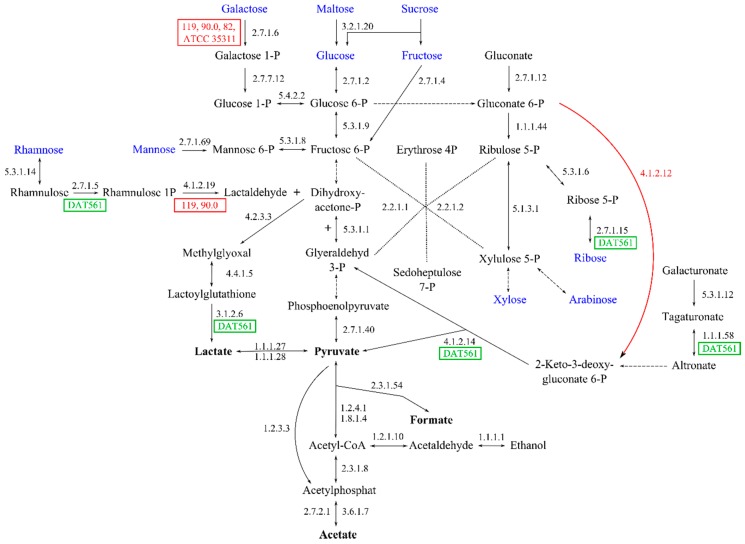
Figure 8.
Genome-based analysis of glycolysis, pentose phosphate pathway, Entner-Doudoroff (ED) pathway, mixed acid fermentation and sugar interconversions (partly). The reactions are schematized (co-factors, co-substrates, CO2-formation are not shown). Dotted arrows indicate a summary of multiple reactions, which were found to be present in all strains. Gene products are visualized via EC numbers for the catalyzing enzymes. Green blocks indicate strain-specific reactions. Red blocks display all strains missing the respective enzyme. Sugars present in honey and degraded pectin backbones (Figure S3) are visualized in blue. Pyruvate and putative end products are shown in bold. All strains lack a pyruvate-decarboxylase (EC 4.1.1.1), which is part of ethanol fermentation. Additionally, all strains lack a phosphogluconate dehydrogenase (red arrow, EC 4.1.2.12), which is part of the Entner-Doudoroff-pathway.