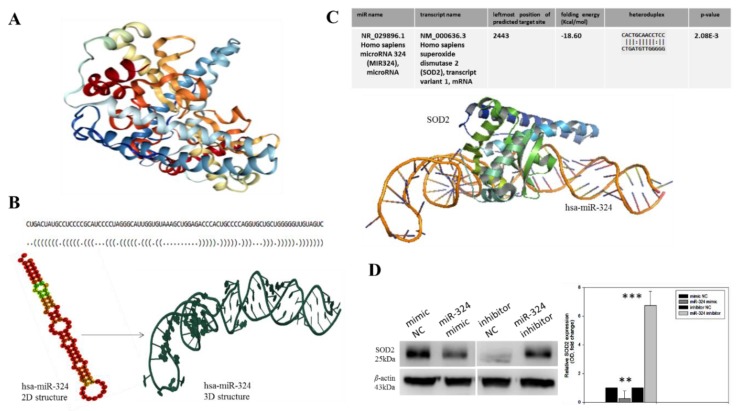
Figure 2.
SOD2 interacts with, and is modulated by hsa-miR-324. (A) Crystal structure of the human manganese superoxide dismutase, PDB: 2ADQ. (B) The optimal secondary structure in dot-bracket notation with a minimum free energy (MFE) of −35.90 kcal/mol, (upper panel). The RNAfold-generated MFE secondary folding structure pattern of hsa-miR-324 (lower panel, left). The hsa-miR-342 tertiary structure generated based on the secondary structure in dot-bracket notation (lower panel, right). (C) Molecular docking showing the direct interaction between SOD2 and hsa-miR-324 with a shape complementarity/docking score of 14,236, Atomic contact energy (ACE) of −33.40 and an approximate SOD2-hsa-miR-324 complex interface area of 1896.30, using a root mean square distance (RMSD) of 9 for the final docking. The 3D transformation data, consisting of three rotational angles (38.44°, 20.02°, 216.62°) and three translational parameters (−0.02, 0.94, −1.95) applied on the ligand molecule, hsa-miR-324. (D) Representative Western blot image (left panel) and quantitative graphical representation (right panel) of the effect of variations in hsa-miR-324 expression on the expression level of SOD2 protein. β-actin served as loading control. NC, negative control; OD, optical density; * p < 0.05, ** p < 0.01, *** p < 0.001.