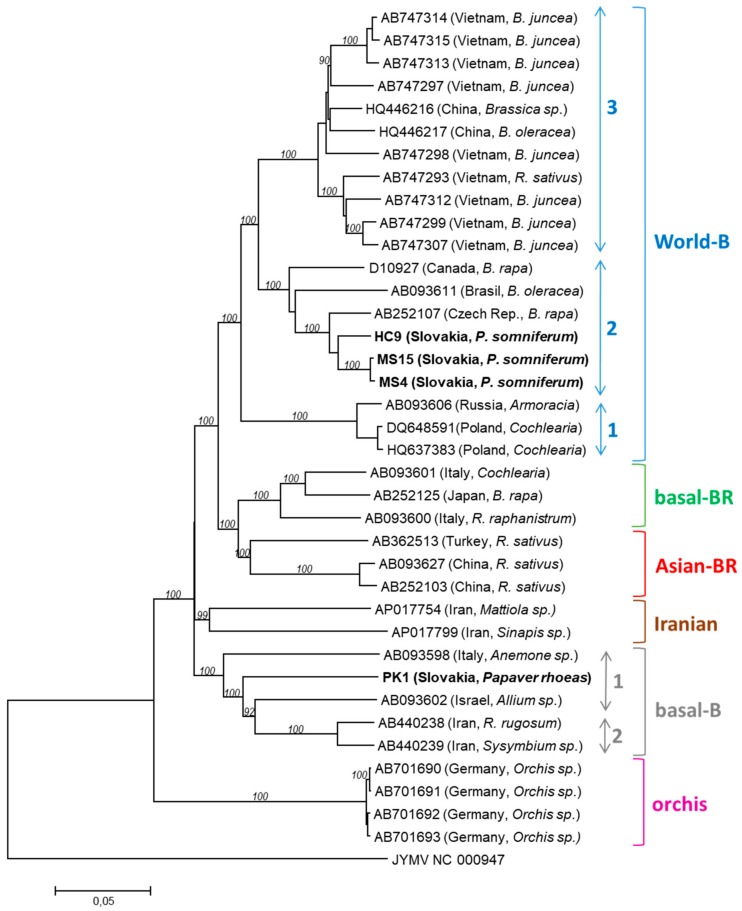
Figure 1.
Neighbour-joining tree inferred from the nearly complete TuMV genomes including those determined in this work (in bold). Non-recombinant database sequences were used (according to [14,24]), identified by acc. number, original host and geographical location. Bootstrap percentages (>70%) based on 1000 pseudoreplicates are indicated at a given node. The scale bar indicates 0.05 substitutions per site. The genomic sequence of Japanese yam mosaic virus (JYMV) was used as an outgroup.