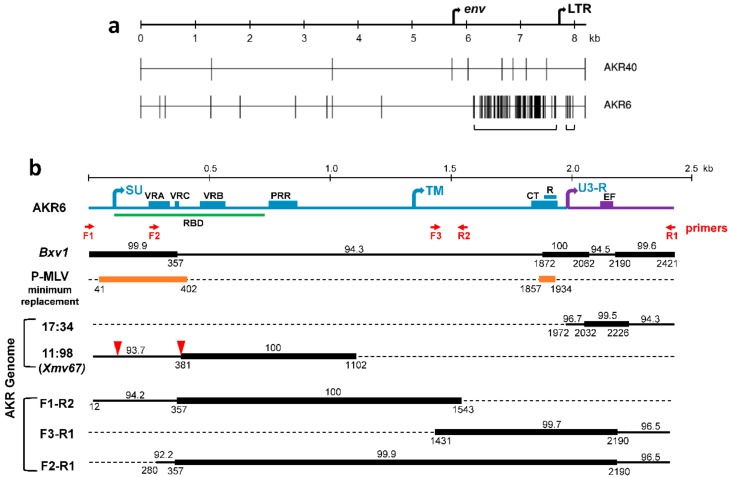
Figure 1.
Comparative analysis of AKR6 and AKR40 X-MLVs. (a) Highlighter plots use vertical lines to identify nucleotide mismatches relative to the Bxv1 Xmv. Brackets indicate two substituted segments in AKR6 relative to Bxv1. (b) Sequence relationships between AKR6 and related endogenous retroviruses (ERVs) and PCR-amplified segments from AKR mice. The env-U3-R segment of the viral genome diagramed at the top notes the locations of the three variable domains (VRA, VRB, and VRC), the proline rich region (PRR) and the receptor binding domain (RBD) within the surface (SU) domain of Env, the cytoplasmic tail (CT) and R-peptide (R) in the transmembrane (TM) domain, and the enhancer framework (EF) within U3. Six black lines show segmental sequence comparisons to AKR6 for Bxv1 and 5 AKR-derived AKR6-related sequences, two from the AKR genome representing a solo long terminal repeat (LTR) and a partial env with two duplications associated with sequencing gaps (red triangles), and three PCR products from AKR amplified using the indicated primers (in red). The orange line positions the two replacements common to the P-MLVs. The numbers above each line show % identity to AKR6, and the numbers below the line identify the start and stop positions of each segment relative to AKR6. Identity > 99% is shown with a thick line.