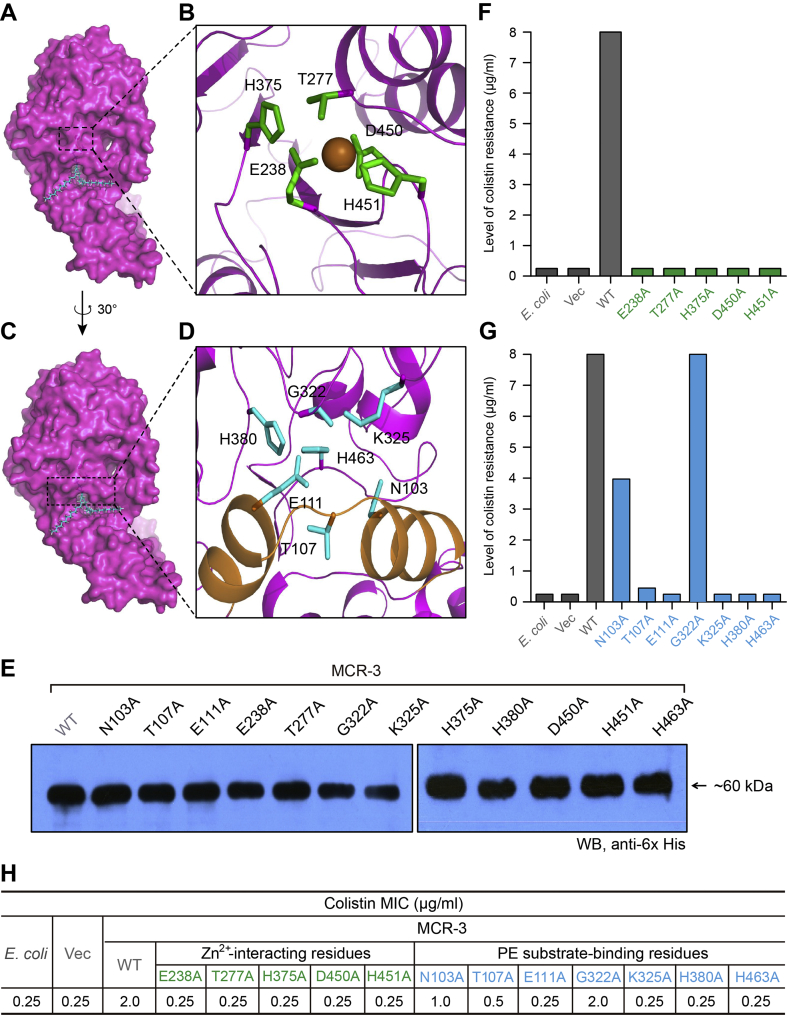
Fig. 6.
Structure-guided functional dissection of MCR-3 colistin resistance.
A. Surface structure of MCR-3 with the cavity required for its PE lipid substrate entry.
B. An enlarged illustration for a five residues-containing, Zn2+-binding motif.
The five residues in Zn2+-binding motif of MCR-3 refers to E238, T277, H375, D450 and H451, respectively.
C. Surface architecture of MCR-3 in counter-clockwise rotation (30 °).
D. An enlarged view of the seven residues-containing motif that is involved in binding of MCR-3 to the PE lipid substrate.
The seven residues denote N103, T107, E111, G322, K325, H380 and H463, respectively.
E. Western blot analyses of the expression of MCR-3 and its 12 point-mutants in E. coli.
F. Structure-guided site-directed mutagenesis analyses for the Zn2+-binding motif of MCR-3 using the colistin susceptibility assays.
G. Functional mapping of the PE-interactive residues of MCR-3 in the context of colistin resistance.
H. The measurement of colistin MIC of the E. coli strains carrying the wild-type mcr-3 (and/or its point-mutants).
Level of colistin resistance was measured using the LBA plates. A representative result is given from no less than three independent trials.
In terms of level of colistin resistance, MCR-3 is almost identical to that of EptA, but only half of that seen with MCR-1.