Abstract
Purpose
Diffusion‐weighted Magnetic Resonance Imaging (DWI) has been shown to be a powerful tool for cancer detection with high tumor‐to‐tissue contrast. This study aims to investigate the feasibility of developing a four‐dimensional DWI technique (4D‐DWI) for imaging respiratory motion for radiation therapy applications.
Materials/Methods
Image acquisition was performed by repeatedly imaging a volume of interest (VOI) using an interleaved multislice single‐shot echo‐planar imaging (EPI) 2D‐DWI sequence in the axial plane. Each 2D‐DWI image was acquired with an intermediately low b‐value (b = 500 s/mm2) and with diffusion‐encoding gradients in x, y, and z diffusion directions. Respiratory motion was simultaneously recorded using a respiratory bellow, and the synchronized respiratory signal was used to retrospectively sort the 2D images to generate 4D‐DWI. Cine MRI using steady‐state free precession was also acquired as a motion reference. As a preliminary feasibility study, this technique was implemented on a 4D digital human phantom (XCAT) with a simulated pancreas tumor. The respiratory motion of the phantom was controlled by regular sinusoidal motion profile. 4D‐DWI tumor motion trajectories were extracted and compared with the input breathing curve. The mean absolute amplitude differences (D) were calculated in superior–inferior (SI) direction and anterior–posterior (AP) direction. The technique was then evaluated on two healthy volunteers. Finally, the effects of 4D‐DWI on apparent diffusion coefficient (ADC) measurements were investigated for hypothetical heterogeneous tumors via simulations.
Results
Tumor trajectories extracted from XCAT 4D‐DWI were consistent with the input signal: the average D value was 1.9 mm (SI) and 0.4 mm (AP). The average D value was 2.6 mm (SI) and 1.7 mm (AP) for the two healthy volunteers.
Conclusion
A 4D‐DWI technique has been developed and evaluated on digital phantom and human subjects. 4D‐DWI can lead to more accurate respiratory motion measurement. This has a great potential to improve the visualization and delineation of cancer tumors for radiotherapy.
Keywords: 4D‐MRI, DWI, IGRT, motion management, retrospective phase sorting
1. Introduction
Four‐Dimensional Magnetic Resonance Imaging (4D‐MRI) is an emerging tool in radiation therapy (RT) to image respiratory motion1, 2, 3, 4 in the abdomen with superior soft tissue contrast and no ionizing radiation hazard comparing to Four‐Dimensional Computer Tomography (4D‐CT).5, 6, 7, 8 Recently, a number of groups have proposed different 4D‐MRI techniques, which can be grouped into two categories: prospective 4D‐MRI techniques and retrospective 4D‐MRI techniques.9
Prospective 4D‐MRI techniques can be further categorized into two types based on its raw image acquisition method: real‐time 3D MRI acquisition and fast 2D MRI acquisition with respiratory gating. Blackall et al. implemented a T1‐weighted 3D real‐time MRI using a Fast Field Echo‐Echo Planar Imaging (FFE‐EPI) sequence with sensitivity encoding (SENSE) parallel acceleration.10 It achieved a high frame rate (330 ms/volume) and spatial resolution (1.8 × 1.8 × 7 mm). However, significant compromises in image quality11 have to be made to achieve real‐time imaging, resulting in an inadequate image quality for RT application. Hu et al. developed a prospective T2‐weighted 4D‐MRI technique with a respiratory amplitude‐based triggering system to gate 2D MRI image acquisition.12 In addition, Akçakaya et al. developed a T1‐weighted 4D‐MRI k‐space‐dependent respiratory gating technique. With this technique, a respiratory navigator is used as the surrogate to gate the acquisition of the k‐space center data.13 Both techniques acquire data only when respiratory motion reaches preset motion amplitude using prospective gating.
Retrospective 4D‐MRI continuously acquires images at all respiratory phases and all slice locations, then retrospectively sorts them by respiratory phases calculated from synchronized motion signal.9, 14, 15, 16 Cai et al. proposed a T2/T1‐weighted 4D‐MRI phase‐sorting technique that used body area (BA) extracting from images as the respiratory surrogate for sorting.9, 17 Tryggestad et al. and Liu et al. separately developed a T2‐weighted 4D‐MRI phase‐sorting technique for sequential MRI image acquisition mode with external respiratory surrogate Physiologic Monitoring Unit (PMU) system.15, 18 In addition, Liu et al. also implemented a retrospective reordering of k‐space based on respiratory phase to generate T2‐weighted 4D‐MRI.16 In summary, 4D‐MRI techniques that have been reported so far have utilized both prospective and retrospective approaches and have utilized different contrast mechanisms (T1‐, T2/T1‐, and T2‐weighted). These techniques are promising in achieving high spatial and/or temporal resolutions. However, from the limited number of patient studies of these techniques,9, 17 it has been seen that current 4D‐MRI techniques suffer from insufficient or inconsistent tumor‐to‐tissue contrast for RT applications.
It is well known that diffusion‐weighted MR imaging (DWI) offers anatomical and functional information with excellent tumor‐tissue contrast.19 It is an important auxiliary MRI weighting showing great promise for detection, cancer diagnosis, tumor target delineation, and cancer treatment response assessment.20 Owing to its high tumor‐to‐tissue contrast and the advantage of not requiring exogenous contrast medium, it has been increasingly used in various clinical applications. DWI of the brain is a well‐established and reliable method for identifying many neurologic cancers.21, 22 For abdominal cancers, DWI has also been shown to have superior tumor‐to‐tissue contrast as compared to CT, T1‐weighted, and T2‐weighted MR images for cancer detection, as shown in Fig. 1. High contrast of DWI leads to high sensitivity and specificity of DWI for detecting abdominal cancer.23, 24 For example, Kartalis et al. conducted a patient study to evaluate the performance of DWI in diagnosis of pancreas cancer. The study shows that DWI has high sensitivity, specificity, and accuracy (92%, 97%, 96%) in diagnosis of pancreatic cancer, similar to comprehensive MRI using multiple sequences (T1‐, T2‐, and contrast‐enhanced perfusion MRI).23 In addition, as a functional imaging method, DWI can be used for assessing treatment response and adaptive RT planning.25, 26, 27 However, respiratory motion in the abdomen largely decreases the quality of DWI images. Respiratory motion can cause substantial errors in target delineation and dose delivery in radiation therapy for thoracic and abdominal cancers when the patient's breathing is irregular.28 In recent years, several techniques have been developed to accelerate the image acquisition, resulting in the reduction in motion artifacts, including the developments of parallel imaging,29, 30 compressed sensing31 , and simultaneous multislice acquisition techniques.32, 33, 34 These techniques have been instrumental to extend the applications of DWI to extracranial sites, including the abdomen and pelvis. For example, the introduction of parallel imaging enabled the reduction in the echo train length, and the k‐space filling time, leading to substantially less motion artifacts at image acquisition, thus enabling high‐quality 2D‐DWI images of the body to be obtained. Furthermore, respiratory‐triggered DWI35, 36 and breath‐hold DWI37 have been investigated for motion artifact‐reduced 3D‐DWI. Chen et al. developed a high‐resolution 3D‐DWI for the abdomen using multishot scan strategy and multiplexed sensitivity encoding (MUSE) to correct nonlinear shot‐to‐shot phase variations without using navigator echoes. K‐space data with the same respiratory amplitude were retrospectively combined for 3D reconstruction.38 Although the existing rapid imaging technique and breathing‐hold technique handle the respiratory motion, it does not provide comprehensive respiratory motion information for treatment of abdominal cancers.
Figure 1.
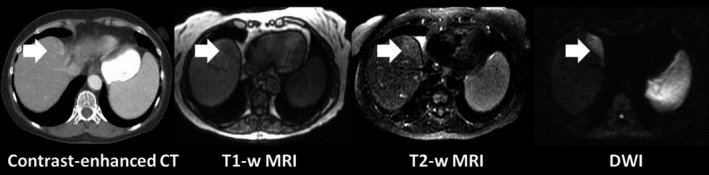
Illustration of difference in tumor contrast in CT and MR images for a pancreatic tumor.
The aim of this study is to investigate the feasibility of a novel 4‐D technique based on diffusion‐weighed MRI (4D‐DWI) for imaging respiratory motion in radiation therapy applications for abdominal cancers, including cancers of pancreas, liver, and many other organs. Compared to other 4D‐MRI techniques, 4D‐DWI potentially offers unique advantages due to its high tumor contrast and capability for functional imaging. In addition to radiotherapy, 4D‐DWI may also have applications for cancer detection and therapeutic response assessment. For example, Apparent Diffusion Coefficient (ADC) maps derived from DWI have been shown to be promising tools for treatment assessment.26 However, their inconsistent reproducibility in the abdomen due to respiratory motion poses a significant challenge for precise measurement of therapeutic changes. The proposed 4D‐DWI technique, with the ability to resolve more accurate tumor information at each respiratory phase, may reduce the motion artifacts and blurring of DWI images. This may subsequently improve ADC measurements. To investigate the effects of 4D‐DWI on ADC measurements, we also conducted computer simulations to compare ADC maps of 4D‐DWI with those from free‐breathing DWI.
2. Methods and materials
2.A. 4D‐DWI technique
The 4D‐DWI technique was achieved by employing a single‐shot echo‐planar imaging (EPI) 2D‐DWI sequence to acquire axial images continuously throughout the breathing cycle under free breathing, and then retrospectively sorting the images based on respiratory phases in three diffusion‐encoding directions separately. 4D‐DWI was then generated by combining the sorted images in different diffusion‐encoding directions. Image acquisition was performed by repeatedly imaging a volume of interest (VOI) using an interleaved multislice single‐shot EPI 2D‐DWI sequence, which is generally available on the major vendor platforms, in the axial plane with free‐breathing. The acquired raw data were exported and processed offline to generate individual 2D‐DWI images for all slices and repetitions without averaging as the input of the retrospective phase‐sorting algorithm.17 Each 2D‐DWI image with an intermediately low b‐value (b = 500 s/mm2) is acquired in x, y, and z diffusion directions. Within one repetition, 2D‐DWI images are acquired in an interleaved fashion with b = 0, and then once for each of the three directions sequentially, that is, the images are acquired with a b‐value of 0 (slice 1, 3, 5…2, 4, 6…), and then b = 500 for the x diffusion direction (slice 1, 3, 5…2, 4, 6…), as well as for the y and z diffusion directions, respectively. Then, the acquisitions are repeated until the entire VOI has been acquired. Respiratory motion is simultaneously recorded along with image acquisition, using a Physiologic Monitoring Unit (PMU) with a pneumatic device (a respiratory bellows) wrapped around the subjects’ upper abdomen as an external respiratory surrogate. The synchronously acquired respiratory signal is used in the retrospective phase‐sorting algorithm17 to generate 4D images using the following four 2D image datasets: the datasets with the b = 0; the datasets with b = 500 in each of the x, y, and z diffusion directions. The first sorted 4D image dataset with b = 0 is labeled as 4D‐DWI (b = 0). The three other sorted 4D image datasets are subsequently combined to reconstruct 4D‐DWI (b = 500 s/mm2). The process is illustrated in Fig. 2. Cine MRI using steady‐state free precession (balanced SSFP)36 is acquired as a reference showing respiratory motion.
Figure 2.
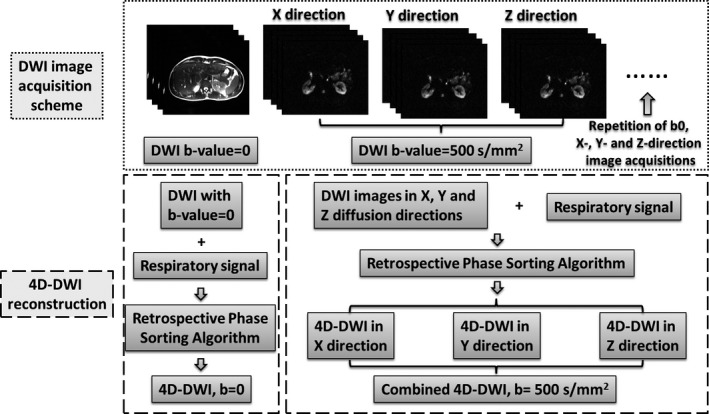
Overall design of the 4D‐DWI technique, including image acquisition scheme, reconstruction process.
2.B. Digital phantom study
The proposed 4D‐DWI technique was first tested via computer simulation using a 4D Digital Extended Cardiac‐Torso (XCAT) human phantom.39, 40, 41 The respiratory motion of the XCAT phantom was controlled by a given regular sinusoidal regular breathing profile (period: 5 s). The peak‐to‐peak motion amplitude of diaphragm in the superior–interior (SI) direction were set to be 3.0 cm and chest wall motion in the anterior–posterior (AP) direction was set to be 1.0 cm. There was no motion in the lateral direction. The XCAT images were generated for abdominal region using the following parameters: in‐plane resolution: 256 × 256; voxel size: 2.5 × 2.5 mm2; slice thickness: 3 mm; one diffusion direction 2D image frame rate: 2.48 Hz. The XCAT phantom was generated in the activity mode for DWI imaging where the signal intensities of the organs and tissues were assigned using values measured from DWI images with b = 500. Image noise was added to the XCAT DWI images to mimic the real DWI images with noisy background. A simulated homogenous spherical tumor of 20 mm in diameter was placed at the head of the pancreas.
The simulation was carried out in the following steps: (a) mimicking the image acquisition of DWI sequence in an interleave image acquisition fashion by extracting axial 2D XCAT images from the 4D XCAT phantom for a VOI. Since the image intensity measurements of the organs and tissues are comparable on the images with diffusion‐encoding gradients in x, y, and z diffusion directions, only the image acquisition process of one direction was simulated. The time of acquisition for each 2D image was recorded; (b) repeating the acquisition process of the VOI described in Step 1 for a number of times. The number of repetitions should be large enough to satisfy 4D sorting data completion condition15 for all four image datasets; (c) synchronizing the image acquisition time of each 2D image with the controlling respiratory motion signal, and acquiring respiratory phase information for each 2D image from the respiratory motion signal; (d) calculating the respiratory phase bin for each extracted 2D XCAT image; and (e) conducting the retrospective phase sorting to generate the simulated 10‐phase 4D‐DWI (b = 500 s/mm2).
The respiratory motion on the simulated 4D‐DWI was quantified by extracting the motion trajectories of the hypothesized tumor in SI and AP directions. They were compared with the input respiratory curve of XCAT. Absolute amplitude difference between the extracted and input respiratory curves was calculated for each phase bin. The mean absolute amplitude difference (D) was calculated to quantitatively evaluate the accuracy of the respiratory motion demonstrated on 4D‐DWI.
2.C. Human study
The 4D‐DWI technique was then tested on two healthy volunteers under a HIPAA‐compliant IRB‐approved study protocol with informed consent. For each subject, DWI images (b‐value = 500 s/mm2) were acquired in the interleaved mode from mid‐thorax to mid‐pelvis. Single‐slice cine MR images were also acquired in the axial, coronal, and sagittal planes for the same field of view to provide reference of region of interest (ROI) motion for comparison. All images were acquired using a 3T MRI system (Tim Trio, Siemens, Erlangen, Germany). Imaging was performed using an anterior six‐channel flexible array coil in combination with 12–16 elements of the table‐mounted spine array. The subjects were positioned head‐first‐supine with arms down without any immobilization device. They were instructed to breathe normally during the scans. Imaging parameters included: repetition time (TR)/echo time (TE), 6900 ms/61.2 ms; field of view (FOV), 380 × 285 mm; flip angle, 160°; slice thickness, 5 mm; matrix, 128 × 96; pixel spacing: 2.97 × 2.97 mm2; bandwidth, 2604 Hz/pixel. Imaging frame rate ranged from 4‐7 f/s. Siemens’ PMU system was used with the bellows wrapped around the abdomen as an external surrogate indicating respiratory motion signal during image acquisition. The respiratory signal was recorded with a sample rate of 50 Hz and synchronized with DWI image acquisition. The PMU was initiated manually by commenting on the computer controlling the MRI scanner. The total image acquisition time was set to be 5–10 min depending on the imaging parameters and reconstruction parameters. The time was optimized to be minimized for each subject but long enough to meet the data sufficient condition.15 Because of the proportional relationship between scanning time needed and the total number of phase bins,15 the total number of phase bins was decreased from 10, as used in XCAT simulation, to six, in order to minimize the total scanning time and avoid it from going above 10 min for human subjects.
The 4D‐DWI was reconstructed using the following steps: (a) acquiring 2D DWI images by repeatedly imaging a VOI using the interleaved multislice single‐shot EPI 2D‐DWI sequence in the axial plane with free‐breathing. 2D DWI images were acquired with b‐values of 0 and 500 s/mm2. For b = 500, 2D DWI images were acquired in x, y, and z directions, respectively. The acquired images were then grouped to four image datasets based on the images’ b‐values and diffusion directions; (b) recording the human subject's respiratory signal simultaneously using the external surrogate PMU with the bellows; (c) synchronizing the image acquisition time of each 2D image with the controlling respiratory motion signal, and calculating the respiratory phase for each 2D image; and (d) conducting the retrospective phase sorting to generate the six‐phase 4D‐DWI (b = 0) and six‐phase 4D‐DWI (b = 500 s/mm2).
Motion trajectories of the upper pole of the right kidney (defined as the ROI) were extracted from 4D‐DWI and compared with those obtained from the single‐slice cine MR images as a reference. The mean absolute amplitude difference (D) was determined for each comparison.
2.D. Effects of 4D‐DWI on ADC measurement
To investigate the effects of 4D‐DWI on ADC measurements, computer simulations were conducted to compare ADC maps derived from 4D‐DWI with those from free‐breathing DWI. DWI of static ROI was served as reference. The simulation was implemented on the XCAT phantom with a simulated tumor in the liver. To demonstrate the influences of 4D sorting on tumor texture demonstration, the liver tumor inserted in XCAT phantom was designed to be a heterogeneous tumor with varies tumor intensities. The background intensity value was set to be the intensity of liver tissue. Other imaging settings and parameters were the same as the previously described XCAT phantom study. Tumor motion was controlled by a regular breathing profile. Using the same image acquisition and reconstruction process, we generated 4D‐DWI images of the heterogeneous tumor for b = 500 s/mm2 and b = 0 s/mm2, respectively. ADC maps were calculated for each respiratory phase bins of 4D‐DWI using the 4D‐DWI (b = 0) and 4D‐DWI (b = 500 s/mm2). ADC maps were calculated using the following formula:
Where S0 is the signal intensity without the diffusion weighting, S1 is the signal with the gradient. b1 and b2 are the two b‐values, which in our case would be 0 and 500 s/mm2.
As a comparison, free‐breathing DWI was also reconstructed using the same image dataset, with the vendor's reconstruction algorithm without applying the retrospective sorting method. The corresponding ADC maps were then generated for a comparison.
Images were analyzed to calculate mean ADC (mADC) and entropy (E) for the tumor region. Mean value of ROI is commonly used for ROI image analysis to briefly evaluate the image intensities of ROI. Entropy is a quantity representing the degree of randomness or disorder of ROI texture on images. It can describe the variation in an ADC histogram such that a large variation in hepatic ADC is associated with larger entropy values. The definition and method to calculate entropy can be found in literature 39 and 40. One mADC value and one E value was calculated for each 4D dataset for 4D‐DWI and each 3D dataset for free‐breathing DWI. The reference of mADC and E values were calculated from the XCAT original static 3D tumor. To simplify the analysis process, only the moving liver tumor region, the ROI, was used for image analysis. Images of the static 3D tumor were used as the reference for image quality.
Furthermore, in order to investigate effects of the breathing pattern variations, the simulation described above was repeated using 10 previously measured liver cancer patients’ breathing profiles to control the respiratory motion of the XCAT phantom. The same image acquisition, 4D‐DWI and free‐breathing DWI reconstruction, and ADC measurements were simulated. The same image analysis was conducted for each breathing profile. Wilcoxon‐Signed Rank test was conducted to compare the mADC values of 4D‐DWI and free‐breathing DWI. The same test was conducted to compare E values of 4D‐DWI and free‐breathing DWI. In addition to Wilcoxon‐Signed Rank test, the root mean squared error (RMSE)40, 41 of the mean ADC value and entropy for 10 patients’ breathing profiles were calculated. The mean ADC value and entropy of the XCAT original static 3D tumor were used as the observation in RMSE calculation.
3. Results
3.A. Digital phantom (XCAT) study
Figure 3(a) shows the simulated 10‐phase 4D‐DWI of the XCAT phantom. The comparison of motion trajectories of the simulated tumor on reconstructed 4D‐DWI and XCAT input respiratory curve shown in Fig. 3(b). Tumor trajectories measured from the simulated XCAT 4D‐DWI were consistent with the input signal: D is 1.88 mm in SI direction and 0.36 mm in AP direction.
Figure 3.
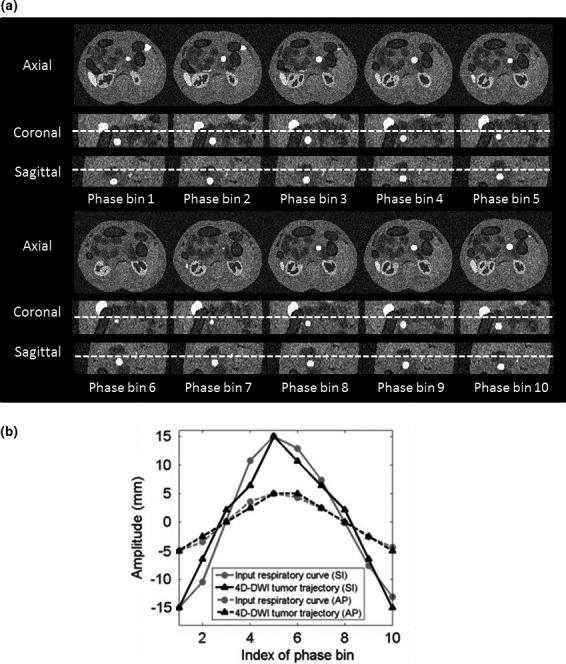
(a) Reconstructed 4D‐DWI simulated with XCAT phantom in axial, sagittal, and coronal view. (b) The difference between tumor trajectories on 4D‐DWI (black) and input respiratory curve (gray) in SI and AP directions.
3.B. Human subject study
Reconstructed 6‐bin 4D‐DWI of the human subjects also demonstrates the respiratory motion. Figure 4(a) shows the 4D‐DWI images of a representative healthy volunteer in axial, coronal, and sagittal view. Figure 4(b) shows the comparison of the ROI motion trajectories extracted from 4D‐DWI images and from the reference cine MRI images. Solid curves are ROI trajectories in SI direction and dashed curves are ROI trajectories in the AP direction. Black curves are trajectories extracted from 4D‐DWI and gray curves are trajectories extracted from reference. It can be seen that the motion trajectories matched generally well between 4D‐DWI and cine MRI. For the two human subjects studied, the mean D value is 2.6 mm in the SI direction and is 1.7 mm in the AP direction.
Figure 4.
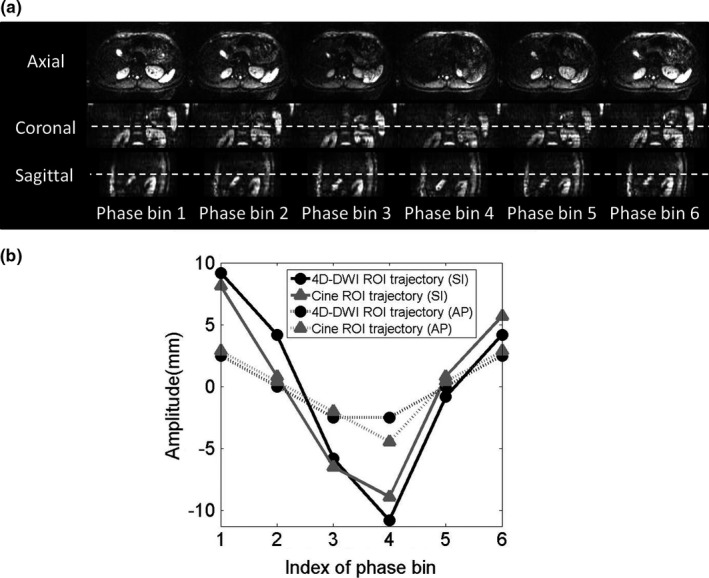
(a) Reconstructed six phase 4D‐DWI images and (b) the motion trajectories of “right kidney (ROI)” for one representative healthy volunteer.
3.C. Effects of 4D‐DWI on ADC measurement
Figure 5 shows the DWI and ADC maps for the XCAT heterogeneous liver tumor with regular respiratory motion. The background intensity is the contrast of liver tissue. To simplify the image quality demonstration and analysis, only the sagittal view of the moving liver tumor region, the ROI, was shown in the figure. The top row shows the comparison of 4D‐DWI, free‐breathing DWI and the original static 3D tumor inserted to the digital phantom. The corresponding ADC maps were shown in the middle row. While the ADC maps measured from free‐breathing DWI provide a motion‐blurred tumor image, those measured from 4D‐DWI demonstrates much clearer tumor texture and motion trajectory information. The bottom row shows the mADC and E values of the tumor region for each ADC image dataset. On average for the six phase bins of 4D‐DWI, mADC value was 2.6 × 10−3 mm2/s. mADC value for free‐breathing condition was 3.9 × 10−3 mm2/s. The reference was 2.6 × 10−3 mm2/s. On average of six phase bins of 4D‐DWI, E value was 0.25. E value for free‐breathing condition was 0.75. The reference was 0.24. The values of both mADC and E were much closer to the reference with the 4D technique.
Figure 5.
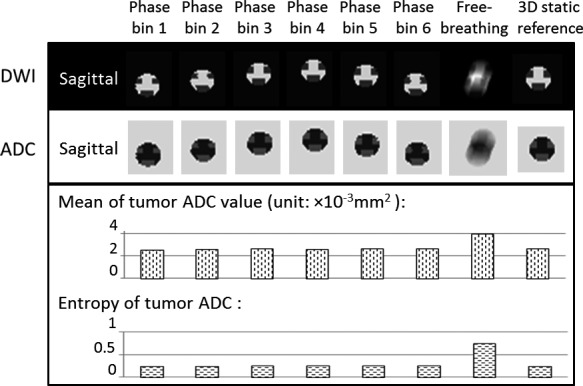
Summary of XCAT phantom 4D‐DWI reconstruction and ADC measurements for the heterogeneous tumor with regular respiratory motion. The top row shows the simulated six‐phase 4D‐DWI images, as compared to the simulated free‐breathing DWI and the referencing static images of the heterogeneous XCAT liver tumor. The corresponding ADC maps were shown in the middle row. To simplify the demonstration, only the sagittal view of the moving liver tumor region was shown. The bottom row shows the mADC and E values of tumor region for each ADC image dataset.
The results of the statistical analysis using 10 patients’ breathing profiles are shown in Fig. 6. On average, the tumor mADC value was 2.7 × 10−3 mm2/s with 4D‐DWI and 4.3 × 10−3 mm2/s with free‐breathing DWI, respectively. The reference was 2.6 × 10−3 mm2/s. The tumor region E was 0.29 with 4D‐DWI and 0.87 with free‐breathing DWI, respectively. The reference was 0.24. The values of both mADC and E were much closer to the reference with the 4D technique. The Wilcoxon‐Signed Rank test shows that, with the 4D‐DWI technique, mADC and E values were significantly different from those calculated from free‐breathing DWI. RMSE of mean ADC values for 4D‐DWI was 0.040 (×0.001 mm2/s); for free‐breathing DWI was 1.634 (×0.001 mm2/s). RMSE of entropy of tumor region for 4D‐DWI was 0.052; for free‐breathing DWI was 0.630. Clearly, RMSE for 4D‐DWI images are much lower than that for free‐breathing DWI.
Figure 6.
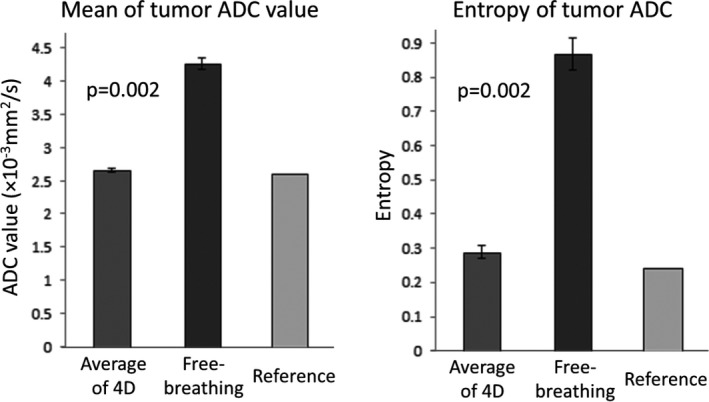
Summary of XCAT phantom mean ADC value and entropy calculated using 10 patients’ breathing profiles. The values measured from the corresponding ADC maps of 4D‐DWI, free‐breathing DWI, and reference were illustrated. Wilcoxon‐Signed Rank test shows that mean ADC value and tumor entropy calculated from 4D‐DWI were significantly more accurate than that from free‐breathing DWI.
4. Discussion
In our study, we developed and evaluated a retrospective 4D‐DWI technique. Feasibility studies on an XCAT digital phantom and in human subjects were conducted. The preliminary results demonstrated that the average respiratory motion of the subjects matches well with motion reference. The 4D‐DWI technique can be applied to liver, pancreas, and many other abdominal regions.
The 4D‐DWI has potential advantages in tumor contrast over other 4D techniques for abdominal cancer imaging. It can lead to a paradigm shift in radiotherapy of abdominal cancer by establishing an entirely new motion management scheme based on 4D‐DWI. This new 4D imaging technique allows more accurate evaluations of tumor respiratory motion, more appropriate design of treatment plans, and more precise delineation of target volumes. It has the potential to be integrated with current and future image‐guided radiotherapy (IGRT) techniques to achieve more accurate patient positioning and targeting. For onboard X‐ray‐based IGRT, we can use the geometric relationships between the target volume contours from the planning 4D‐DWI and other IGRT surrogates, such as stent, fiducials, and cone‐beam CT landmarks, to guide patient positioning. For onboard MRI IGRT, it is also possible to acquire pretreatment verification 4D‐DWI to match with the planning 4D‐DWI for precise targeting.
Despite the wide use of prospective and retrospective sorting techniques in MRI, especially in 4D‐MRI,15, 16, 17, 42 dynamic enhanced contrast (DCE)‐MRI,43 and cardiac imaging,44 to our best knowledge there is no study yet to apply the sorting technique on DWI. This study is the first to demonstrate the feasibility of 4D‐DWI using a retrospective sorting method. Compared to the aforementioned techniques, the implementation of sorting on DWI has different challenges, such as multiple dimensions of diffusion, significant geometric distortions, and large imaging volume (as compared to cardiac imaging). In addition, the DWI sequence used in this study is a work‐in‐progress (WIP) sequence that has much better image quality (better resolutions, less distortion artifacts, higher signal) than the current standard DWI sequence, which made possible the implementation of sorting on DWI.
Compared to previous 4D‐MRI techniques, the 4D‐DWI technique holds two major novelties: (a) the sorting and reconstruction methods of 4D‐DWI are different from those used previously. While most of other 4D‐MRI techniques processed the 2D images to generate 4D‐MRI, the 4D‐DWI technique processes the raw diffusion images with diffusion‐encoding gradients in x, y, and z diffusion directions. The MRI image acquisition and postprocessing pipeline was modified in the MRI scanner to acquire the raw data instead of the reconstructed 2D image. The raw images with three different diffusion directions were processed separately and then combined to generate 4D‐DWI images. This 4D reconstruction process of dealing with raw diffusion image data in different directions and the necessity of synchronizing the raw data with respiratory signal added the complexity of the technique. This is a major alteration of 4D reconstruction technique; (b) the technique is designed to generate 4D images for diffusion MRI. Compared to other 4D‐MRI techniques, 4D‐DWI potentially offers unique advantages due to its high tumor contrast and capability for functional imaging. In addition, the derivative of DWI, ADC maps have been shown to be promising tools for treatment assessment. With all the unique advantages, 4D‐DWI holds great promises for improving the accuracy of tumor volume delineation and treatment response assessment for radiotherapy.
Even though the preliminary results demonstrated the feasibility of the retrospective phase‐sorting 4D‐DWI technique, there are still several problems which need to be clarified: (a) The balance of the temporal resolution, or the frame rate equivalently, and the image quality of the 2D DWI image acquisition. One criterion for 4D‐DWI image acquisition frame rate selection is that the total number of bins should be no larger than the product of average respiratory period and the frame rate of MRI acquisition, that is, NB ≤ fr * T, in which NB is the total number of bins, fr is the frame rate of image acquisition and T is the average respiratory period.15, 18 For example, the typical respiratory period is about 3 s, if six phase bins are selected for 4D reconstruction, frame rate of the 2D image acquisition should be higher than 2 Hz. With the criteria of frame rate higher than 2 Hz, fast 2D‐DWI sequences are required for 4D reconstruction. With the requirement of high frame rate, 2D image quality might be limited or sacrificed. (b) The b‐value for 4D‐DWI needs to be optimized. It is possible that 4D‐DWI with higher b‐values may lose more anatomical details for treatment planning, although higher b‐values maybe preferred due to high tumor contrast. We plan to investigate this by evaluating several b‐values between 500 s/mm2 to 1000 s/mm2 in a future study. (c) In most of the 4D imaging studies, the total number of bins is selected to be 6~10.9, 12, 18 We selected six in our study to minimize the total scanning time. There are pros and cons for selecting a larger number of bins.15, 45 What is the optimized total number of bins is an interesting subject in our future study plan. (d) Furthermore, the quantitative analysis of the effect of 4D‐DWI on time‐resolved ADC maps can be extended in our future study. For the ROIs on time‐resolved ADC maps, more image features, such as tumor homogeneity, tumor sphericity, tumor surface area, tumor contrast, and several other features have the potential to be used for evaluation besides the examples we used in the current study: mean ADC value and Entropy. (e) Focusing on the development of the technique, the number of human subjects in our study is limited. The technique has been successfully implemented, proving that the 4D‐DWI technique is feasible. A study with a large patient number is desirable for a more complete evaluation of the technique. In addition, the statistical analysis on the above image features mentioned in (c) can be conducted once more when human subject data are available. (f) Lastly, the 4D‐DWI image quality evaluation can be expanded to include more current clinical standards of DWI techniques, such as respiratory‐gated DWI, breath‐hold DWI, etc.46 This is a topic of interest and will be included in our future investigations.
5. Conclusion
A novel respiratory‐correlated 4D‐DWI technique has been developed and evaluated on a digital phantom and human subjects. Compared to other 4D MRI techniques, 4D‐DWI is advantageous because of its superior tumor contrast and ability to measure functionality. It holds great promises for improving the accuracy of tumor volume delineation and treatment response assessment for radiotherapy, especially for abdominal cancers.
Conflict of interest
This work is partly supported by funding from NIH (1R21CA165384), a research grant from the Golfers Against Cancer (GAC) Foundation and Siemens Healthcare.
References
- 1. Ozhasoglu C, Murphy MJ. Issues in respiratory motion compensation during external‐beam radiotherapy. Int J Radiat Oncol Biol Phys. 2002;52:1389–1399. [DOI] [PubMed] [Google Scholar]
- 2. Ford EC, Mageras GS, Yorke E, Ling CC. Respiration‐correlated spiral CT: a method of measuring respiratory‐induced anatomic motion for radiation treatment planning. Med Phys. 2003;30:88–97. [DOI] [PubMed] [Google Scholar]
- 3. Dawson LA, Jaffray DA. Advances in image‐guided radiation therapy. J Clin Oncol. 2007;25:938–946. [DOI] [PubMed] [Google Scholar]
- 4. Keall PJ, Mageras GS, Balter JM, et al. The management of respiratory motion in radiation oncology report of AAPM Task Group 76. Med Phy. 2006;33:3874–3900. [DOI] [PubMed] [Google Scholar]
- 5. Vinogradskiy YY, Balter P, Followill DS, Alvarez PE, White RA, Starkschall G. Comparing the accuracy of four‐dimensional photon dose calculations with three‐dimensional calculations using moving and deforming phantoms. Med Phys. 2009;36:5000–5006. [DOI] [PubMed] [Google Scholar]
- 6. Pan T, Lee TY, Rietzel E, Chen GT. 4D‐CT imaging of a volume influenced by respiratory motion on multi‐slice CT. Med Phys. 2004;31:333–334. [DOI] [PubMed] [Google Scholar]
- 7. Tian Z, Jia X, Dong B, Lou Y, Jiang SB. Low‐dose 4DCT reconstruction via temporal nonlocal means. Med Phys. 2011;38:1359–1365. [DOI] [PubMed] [Google Scholar]
- 8. Clark D, Badea A, Liu Y, Johnson GA, Badea CT. Registration‐based segmentation of murine 4D cardiac micro‐CT data using symmetric normalization. Phys Med Biol. 2012;57:6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cai J, Chang Z, Wang Z, Segars WP, Yin F. Four‐dimensional magnetic resonance imaging (4D‐MRI) using image‐based respiratory surrogate: a feasibility study. Med Phys. 2011;38:6384–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blackall JM, Ahmad S, Miquel ME, McClelland JR, Landau DB, Hawkes DJ. MRI‐based measurements of respiratory motion variability and assessment of imaging strategies for radiotherapy planning. Phys Med Biol. 2006;51:4147. [DOI] [PubMed] [Google Scholar]
- 11. Von Siebenthal M, Sz′ekely G, Gamper U, Boesiger P, Lomax A, Cattin Ph. 4D MR imaging of respiratory organ motion and its variability. Phys Med Biol. 2007;52:1547–1564. [DOI] [PubMed] [Google Scholar]
- 12. Hu Y, Caruthers S, Low D, Parikh P, Mutic S. Respiratory amplitude guided 4‐dimensional magnetic resonance imaging. Int J Radiat Oncol Biol Phys. 2013;86:198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Akçakaya M, Gulaka P, Basha T, Ngo L, Manning W, Nezafat R. Free‐breathing phase contrast MRI with near 100% respiratory navigator efficiency using k‐space‐dependent respiratory gating. Magn Reson Med. 2014;71:2172–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cai J, Liu Y, Yin F. Extracting breathing signal using fourier transform from cine magnetic resonance imaging. Cancer Trans Med. 2015;1:16. [Google Scholar]
- 15. Liu Y, Yin F, Czito B, Bashir M, Cai J. T2‐weighted four dimensional magnetic resonance imaging with result‐driven phase sorting. Med Phys. 2015;42:4460–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Y, Yin F, Chen NK, Chu ML, Cai J. Four dimensional magnetic resonance imaging with retrospective k‐space reordering: a feasibility study. Med Phys. 2015;42:534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu Y, Yin F, Chang Z, et al. Investigation of sagittal image acquisition for 4D‐MRI with body area as respiratory surrogate. Med Phys. 2014;41:101902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tryggestad E, Flammang A, Han‐Oh S, et al. Respiration‐based sorting of dynamic MRI to derive representative 4D‐MRI for radiotherapy planning. Med Phys. 2013;40:051909. [DOI] [PubMed] [Google Scholar]
- 19. Taouli B. Diffusion‐weighted MR imaging for diagnosis of liver metastases. Magnetom Flash. 2005;1:44–47. [Google Scholar]
- 20. Koh D, Collins DJ. Diffusion‐weighted MRI in the body: applications and challenges in oncology. AJR Am J Roentgenol. 2007;188:1622–1635. [DOI] [PubMed] [Google Scholar]
- 21. Kono K, Inoue Y, Nakayama K, et al. The role of diffusion‐weighted imaging in patients with brain tumors. AJNR Am J Neuroradiol. 2001;22:1081–1088. [PMC free article] [PubMed] [Google Scholar]
- 22. Chang SC, Lai PH, Chen WL, et al. Diffusion‐weighted MRI features of brain abscess and cystic or necrotic brain tumors: comparison with conventional MRI. Clin Imag. 2002;26:227–236. [DOI] [PubMed] [Google Scholar]
- 23. Kartalis N, Lindholm TL, Aspelin P, Permert J, Albiin N. Diffusion‐weighted magnetic resonance imaging of pancreas tumours. Eur Radiol. 2009;19:1981–1990. [DOI] [PubMed] [Google Scholar]
- 24. Ichikawa T, Erturk SM, Motosugi U, et al. High‐b value diffusion‐weighted MRI for detecting pancreatic adenocarcinoma: preliminary results. AJR Roentgenol. 2007;188:409–414. [DOI] [PubMed] [Google Scholar]
- 25. Tsien C, Cao Y, Chenevert T. Clinical applications for diffusion magnetic resonance imaging in radiotherapy. Semin Radiat Oncol. 2014;24:218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Afaq A, Andreou A, Koh DM. Diffusion‐weighted magnetic resonance imaging for tumour response assessment: why, when and how? Cancer Imag. 2010;10:S179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haack S, Pedersen EM, Jespersen SN, Kallehauge JF, Lindegaard JC, Tanderup K. Apparent diffusion coefficients in GEC ESTRO target volumes for image guided adaptive brachytherapy of locally advanced cervical cancer. Acta Oncol. 2010;49:978–983. [DOI] [PubMed] [Google Scholar]
- 28. Hugo GD, Rosu M. Advances in 4D radiation therapy for managing respiration: part I‐4D imaging. Z Med Phys. 2012;22:258–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42:952–962. [PubMed] [Google Scholar]
- 30. Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med. 2002;47:1202–1210. [DOI] [PubMed] [Google Scholar]
- 31. Lustig M, Donoho D, Pauly JM. Sparse MRI: the application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58:1182–1195. [DOI] [PubMed] [Google Scholar]
- 32. Breuer FA, Blaimer M, Heidemann RM, Mueller MF, Griswold MA, Jakob PM. Controlled aliasing in parallel imaging results in higher acceleration (CAIPIRINHA) for multi‐slice imaging. Magn Reson Med. 2005;53:684–691. [DOI] [PubMed] [Google Scholar]
- 33. Moeller S, Yacoub E, Olman CA, et al. Multiband multislice GE‐EPI at 7 tesla, with 16‐fold acceleration using partial parallel imaging with application to high spatial and temporal whole‐brain fMRI. Magn Reson Med. 2010;63:1144–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feinberg DA, Setsompop K. Ultra‐fast MRI of the human brain with simultaneous multi‐slice imaging. J Magn Reson. 2013;229:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bruegel M, Holzapfel K, Gaa J, et al. Characterization of focal liver lesions by ADC measurements using a respiratory triggered diffusion‐weighted single‐shot echo‐planar MR imaging technique. Eur Radiol. 2008;18:477–485. [DOI] [PubMed] [Google Scholar]
- 36. Gourtsoyianni S, Papanikolaou N, Yarmenitis S, Maris T, Karantanas A, Gourtsoyiannis N. Respiratory gated diffusion‐weighted imaging of the liver: value of apparent diffusion coefficient measurements in the differentiation between most commonly encountered benign and malignant focal liver lesions. Eur Radiol. 2008;18:486–492. [DOI] [PubMed] [Google Scholar]
- 37. Kandpal H, Sharma R, Madhusudhan KS, Kapoor KS. Respiratory‐triggered versus breath‐hold diffusion‐weighted MRI of liver lesions: comparison of image quality and apparent diffusion coefficient values. AJR Am J Roentgenol. 2009;192:915–922. [DOI] [PubMed] [Google Scholar]
- 38. Chen NK, Guidon A, Chang HC, Song AW. A robust multi‐shot scan strategy for high‐resolution diffusion weighted MRI enabled by multiplexed sensitivity‐encoding (MUSE). NeuroImage. 2013;72:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Segars WP, Sturgeon G, Mendonca S, Grimes J, Tsui BMW. 4D XCAT phantom for multimodality imaging research. Med Phys. 2010;37:4902–4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Segars WP, Tsui BMW. MCAT to XCAT: the Evolution of 4‐D computerised phantoms for imaging research. Proc IEEE. 2009;97:1954–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Segars WP, Mahesh M, Beck TJ, Frey EC, Tsui BMW. Realistic CT simulation using the 4D XCAT phantom. Med Phys. 2008;35:3800–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paganelli C, Summers P, Bellomi M, Baroni G, Riboldi M. Liver 4DMRI: a retrospective image‐based sorting method. Med Phys. 2015;42:4814–4821. [DOI] [PubMed] [Google Scholar]
- 43. McGee KP, Hu Y, Tryggestad E, et al. MRI in radiation oncology: underserved needs. Magn Reson Med. 2016;75:11–14. [DOI] [PubMed] [Google Scholar]
- 44. Stankovic Z, Allen BD, Garcia J, Jarvis KB, Markl M. 4D flow imaging with MRI. Cardiovasc Diagn Ther. 2014;4:173–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park SJ, Ionascu D, Killoran J, et al. Evaluation of the combined effects of target size, respiratory motion and background activity on 3D and 4D PET/CT images. Phys Med Biol. 2008;53:3661. [DOI] [PubMed] [Google Scholar]
- 46. Kwee TC, Takahara T, Koh DM, Nievelstein RA, Luijten PR. Comparison and reproducibility of ADC measurements in breathhold, respiratory triggered, and free‐breathing diffusion‐weighted MR imaging of the liver. J Magn Reson Imaging. 2008;28:1141–1148. [DOI] [PubMed] [Google Scholar]


