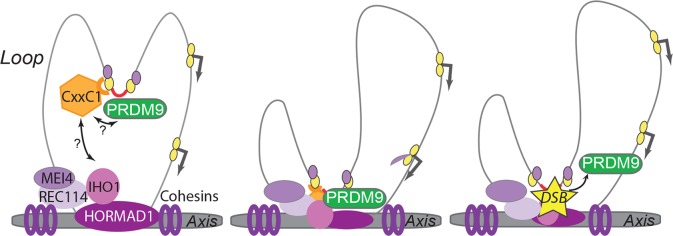
Fig 6. Model of PRDM9 binding dynamics in mouse spermatocytes.
In early meiotic prophase, when PRDM9 is expressed, chromosomes are organized in a characteristic loop axis structure, shaped by cohesins and other proteins. Some essential components of meiotic DSB formation, such as HORMAD1, MEI4, REC114, and IHO1, are located on the axis where they form discrete foci. PRDM9 binds via its zinc finger domain to DNA motifs (red) that define meiotic recombination hotspots and that are likely to be located in chromatin loops (only one sister chromatid is represented). PRDM9 modifies the surrounding nucleosomes by catalysing H3K4me3 (yellow) and H3K36me3 (purple) deposition (left panel). Then, a reader or adaptor protein mediates PRDM9 interaction with the DNA DSB formation machinery located on the axis. This reader could be CXXC1, a H3K4me3 reader that interacts with both PRDM9 and IHO1 (central panel). Therefore, PRDM9 can also indirectly interact with DNA sequences near or on the axis. SPO11 (the protein carrying the catalytic activity for meiotic DSB formation) may be recruited at PRDM9-binding sites before or after the loop axis interaction. Upon DSB formation, PRDM9 could be displaced and could interact with other sites, such as transcription start sites, possibly through interaction with other unknown factors (right panel). DSB, DNA double strand break; H3K4me3, histone H3 lysine4 trimethyl.