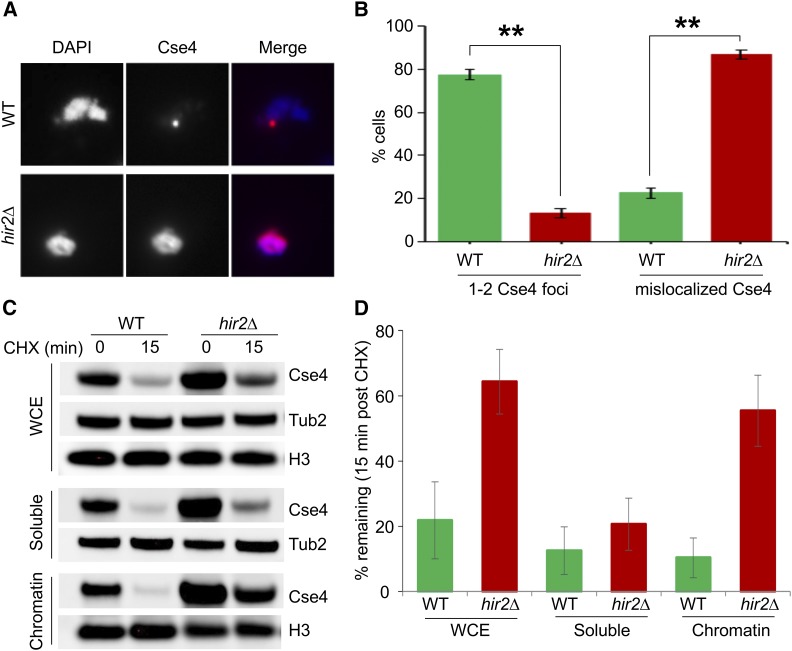
Figure 5.
Cse4 mislocalized to noncentromeric regions in hir2∆ strains. (A) Cse4 is mislocalized in hir2∆ strains. Chromosome spreads were prepared from WT and hir2∆ strains with GALCSE4HA (pMB1458) grown in SC-Ura containing 2% raffinose. Expression of Cse4 was induced for 1 hr by adding 2% galactose. Chromosome spreads were probed with α-HA (16B12; Covance) primary antibodies followed by Cy3 conjugated Goat α-mouse secondary antibodies for Cse4 detection. Nuclear mass was visualized by DAPI staining. (B) Quantification of Cse4 mislocalization in hir2∆ strains. Percentage of cells from chromosome spreads in (A) that exhibit either 1–2 Cse4-foci or dispersed Cse4 signals in WT or hir2∆ strains. (C and D) Cse4 is stable in the chromatin fraction from hir2∆ strains. The levels and stability of Cse4 in whole cell extract (WCE), soluble, and chromatin fractions were determined by Western blot analysis using α-HA antibody. Expression of Cse4 was induced in WT or hir2∆ strains by growth in galactose-containing medium for 30 min followed by treatment with CHX (10 μg/ml) for 15 min. Tub2 and histone H3 were loading controls. (D) Quantification of Cse4 levels from (C). The percentage of Cse4 remaining after CHX treatment (15 min) is calculated taking the t0 value as 100%. The error bar represents the average deviation from the mean of two independent biological repeats.