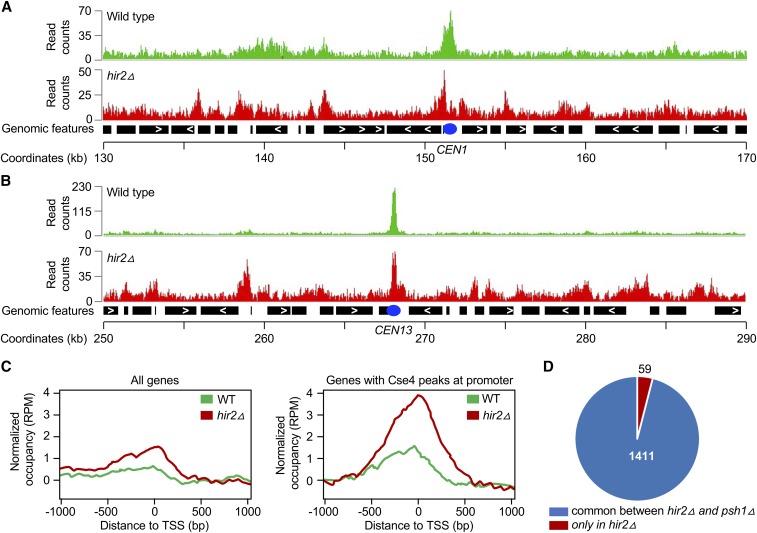
Figure 6.
Cse4 is mislocalized to noncentromeric regions in hir2∆ strains. ChIP was performed using chromatin prepared from WT and hir2∆ strains carrying GALCSE4HA (pMB1458) after growth for 6 hr in SC-Ura with galactose and raffinose (2% each). Immunoprecipitation was with α-HA agarose as described in Materials and Methods. Input and immunoprecipitated samples were used for ChIP-sequencing as described (Grøntved et al. 2015). (A and B) Cse4 is mislocalized to noncentromeric regions in a hir2∆ strain. A representative region of the Cse4 ChIP-seq enrichment from WT and hir2∆ strains is shown from the chromosome 1 region flanking CEN (between 130,000 and 170,000 bp). Peaks of Cse4 after normalization and input subtraction are shown. (B) A representative region of the Cse4 ChIP-seq enrichment from WT and hir2∆ strains is shown from the chromosome 13 region flanking CEN (between 250,000 and 290,000 bp). Peaks of Cse4 after normalization input subtraction are shown. (C) Mislocalization of overexpressed Cse4 is higher in promoter regions. Average Cse4 ChIP-seq coverage from WT and hir2∆ strains calculated from 1000 bp upstream and downstream of transcription start sites (TSS) for all genes or for regions where Cse4 was found to be enriched at promoters. (D) Comparative analysis of genomic regions for Cse4 mislocalization between hir2∆ and psh1∆ strains. Pie chart denotes the genomic regions associated with mislocalized Cse4 common between hir2∆ strain from our study and psh1∆ strain identified previously (Hildebrand and Biggins 2016).