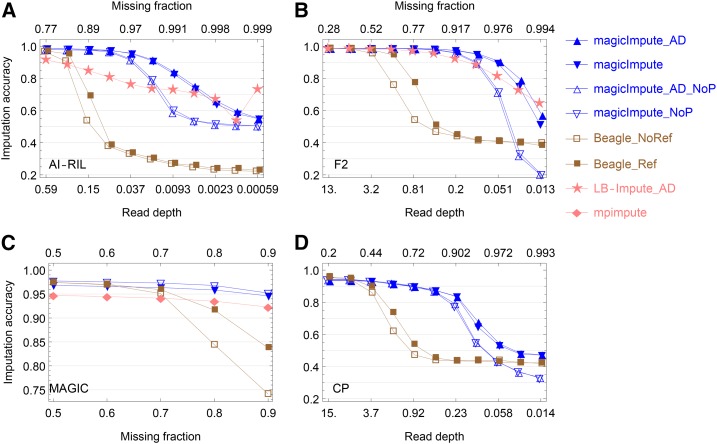
Figure 5.
The accuracy of imputing offspring genotypes from real data. (A–D) The results for the AI-RIL, the F2, the MAGIC, and the CP, respectively, are shown. In the figure legend on the right side, “_AD” denotes that the input data are allelic depths rather than called genotypes, “_NoP” denotes that the first two founders’ genotypes are not available, and “_Ref” and “_NoRef” denotes whether Beagle uses founder haplotypes as reference panels or not. Allelic depth data are not available for the MAGIC. The extreme large missing fraction or low read depth shows how genotype imputation approaches random imputation with decreasing amount of the input data. In (A) the large variation of imputation accuracy of LB-Impute at low read depths is due to the corresponding imputation fraction being close to 0 (Figure S8B).