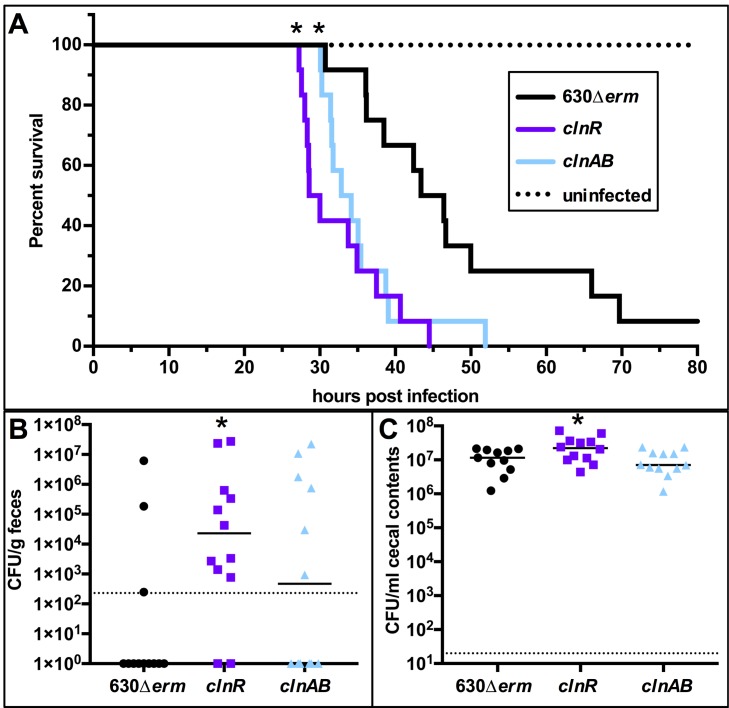
Fig 6. clnR and clnAB mutants are more virulent in a hamster model of infection.
Syrian golden hamsters were inoculated with approximately 5000 spores of strain 630Δerm (n = 12), clnR (MC885; n = 12), or clnAB (MC935; n = 12). A) Kaplan-Meier survival curve depicting time to morbidity. Mean times to morbidity were: 630Δerm 46.0 ± 12.2 (n = 11); clnR 32.5 ± 5.8 (n = 12); clnA 35.2 ± 6.1 (n = 12). * indicates p ≤ 0.01 by log-rank test. B) Total C. difficile CFU recovered from fecal samples collected at 12 h.p.i. Dotted line demarcates lowest limit of detection. Solid black line marks the median. Fisher’s exact test compared the number of animals without and with detectable CFU compared to 630Δerm (* indicates p < 0.05). C) Total C. difficile CFU recovered from cecal contents collected post-mortem. Dotted line demarcates limit of detection. Solid black line marks the median. Numbers of CFU are compared to 630Δerm by one-way ANOVA with Dunnett’s multiple comparisons test (* indicates p < 0.05).