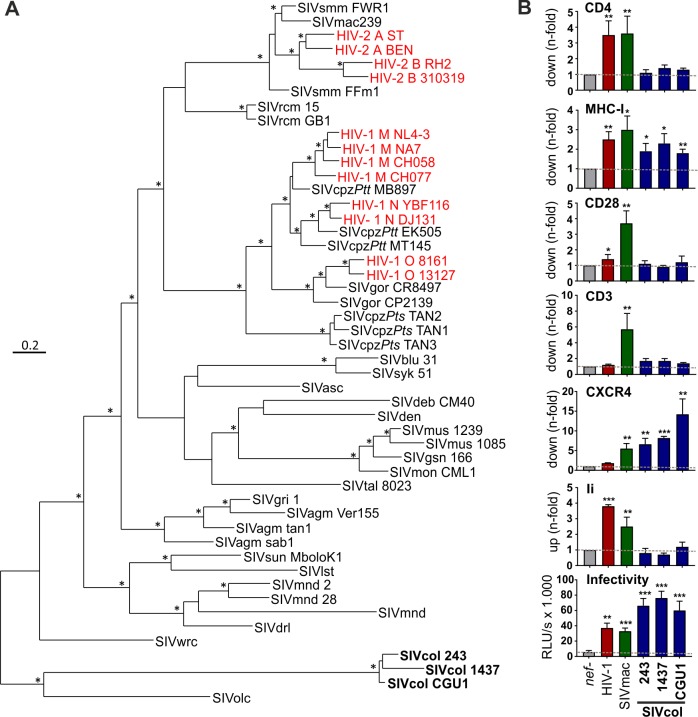
Fig 1. Phylogenetic position and functional activity of SIVcol Nef proteins.
(A) Phylogenetic relationship of Nef amino acid sequences from different primate lentiviruses. Stars on branches indicate ≥90 percentage estimated posterior probabilities. The length of a branch indicates the phylogenetic distance to its origin. (B) Modulation of various receptors and viral infectivity enhancement by SIVcol Nefs. To measure receptor modulation, human PBMCs, Jurkat (for CXCR4) or THP-1 cells (for Ii) were transduced with HIV-1 NL4-3 IRES-eGFP constructs containing the indicated nef alleles. The mean channel numbers of red fluorescence obtained for cells transfected with the nef-defective HIV-1 construct were divided by the corresponding numbers obtained for cells infected with viral constructs coexpressing Nef and eGFP to calculate n-fold down- or up-modulation, respectively. The lowest panel shows the β-galactosidase activity (RLU/s) obtained after infection of P4-CCR5 cells with virus stocks containing 5 ng p24 antigen produced by transfection of HEK293T cells. Results show mean values (+SEM) derived from six to eight measurements in at least two independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.