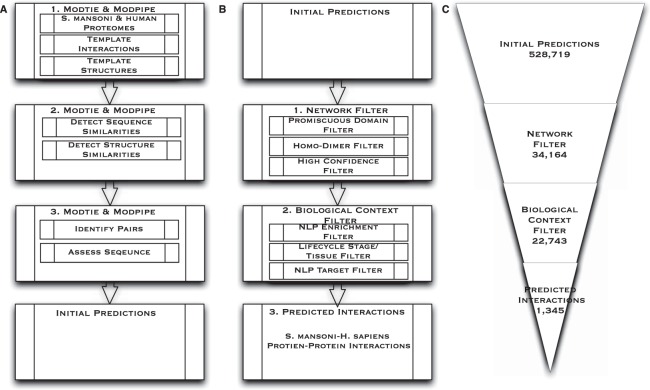
Figure 1.
Prediction Framework. (A) Modtie & ModPipe protocol for detecting sequence and structure similarity: 1. The protocol begins with the set of human and S. mansoni proteins. 2. Sequence matching procedures are then used to identify similarities between the proteins and proteins with known structure or interactors. 3. A structure-based statistical potential assessment, or a sequence similarity score in the absence of structure, is then used to identify pairs with similarity to known complexes, assess the basis for a putative interaction, predict interacting partners and yield the Initial Predictions. (B) Initial Predictions: 1. Network Filter: Promiscuous Dimers, homo-dimer complexes, and high confidence interactions are then extracted from the initial set of predictions. 2. The Biological Context Filter is applied to the remaining set of predictions, weighting and ranking NLP (natural language processing) enriched predictions, isolating life cycle stage and tissues interactions between S. mansoni and humans, and application of the Targeted Filter. 3. Predicted Interactions are an output of the Biological Context Filter. (C) This Illustrates the numerical reduction in interactions obtained after each step. The framework reduces the number of potential S. mansoni-human protein interactions by about three orders of magnitude as shown here and in Tables 1–3.