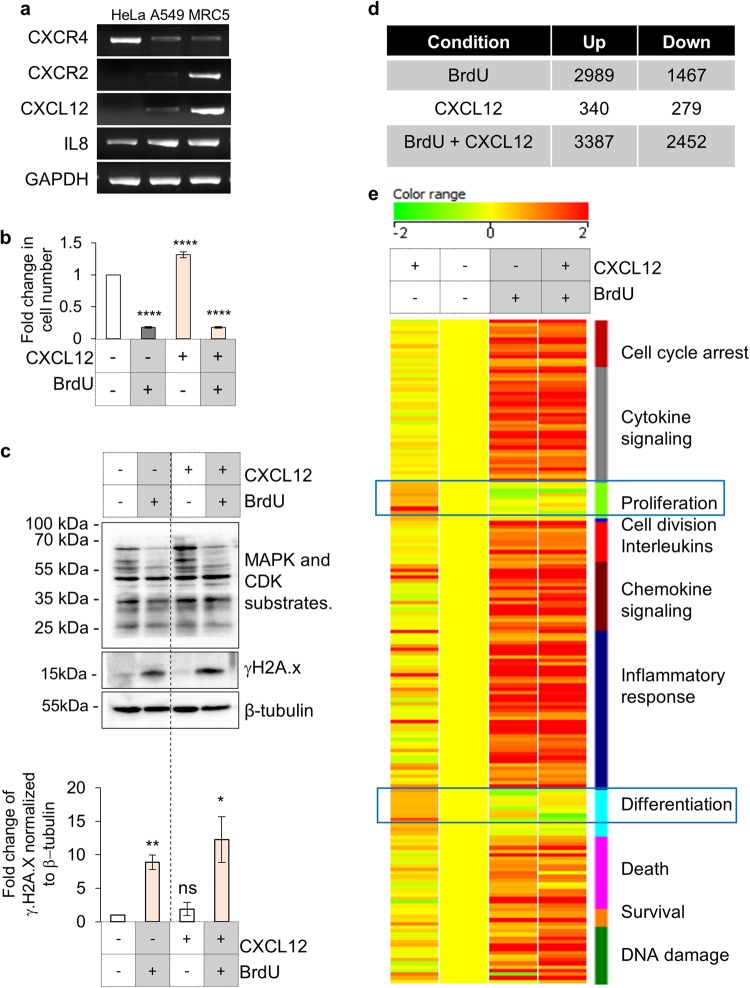
Fig. 3.
Analysis of effects of CXCR4 activation in damaged cells. a Gene expression analysis to evaluate transcription of CXCR4 and CXCR2 and respective ligands by PCR. Expression analysis of mentioned genes was performed in HeLa, A549 and MRC5 cells (Table S2). b Effect of CXCL12-CXCR4 axis on cellular proliferation. HeLa cells, with or without CXCL12 (200 ng/ml) or BrdU were counted after 72 h of treatment. The Y-axis represents fold change of proliferative index wrt undamaged unstimulated cells. Results are mean ± s.e.m. ****p ≤ 0.0001 (Student’s t-test; n > 10). c Analysis for γH2A.x and MAPK/CDK substrate phosphorylation on activation of CXCR4 receptor. HeLa were treated with CXCL12 (200 ng/ml) in presence or absence of 100 µM BrdU for 48 h and the levels of MAPK-CDK substrates (top), phospho-H2Ax (middle panel) and β-tubulin (bottom) were analyzed by western blotting. Quantitation of γH2Ax levels are shown in the graph (n = 3). d Microarray analysis of gene expression changes in response to CXCL12 treatment. Table shows number of gene up or downregulated in comparison to untreated HeLa cells in presence of BrdU, CXCL12 or BrdU with CXCL12 after 48 h (n = 2). e Heat map of functional clusters in microarray data. Genes differentially regulated in presence of BrdU or CXCL12 alone or with BrdU along with CXCL12 (as indicated) were clustered into indicated families and heat map was generated based on fold changes (n = 2)