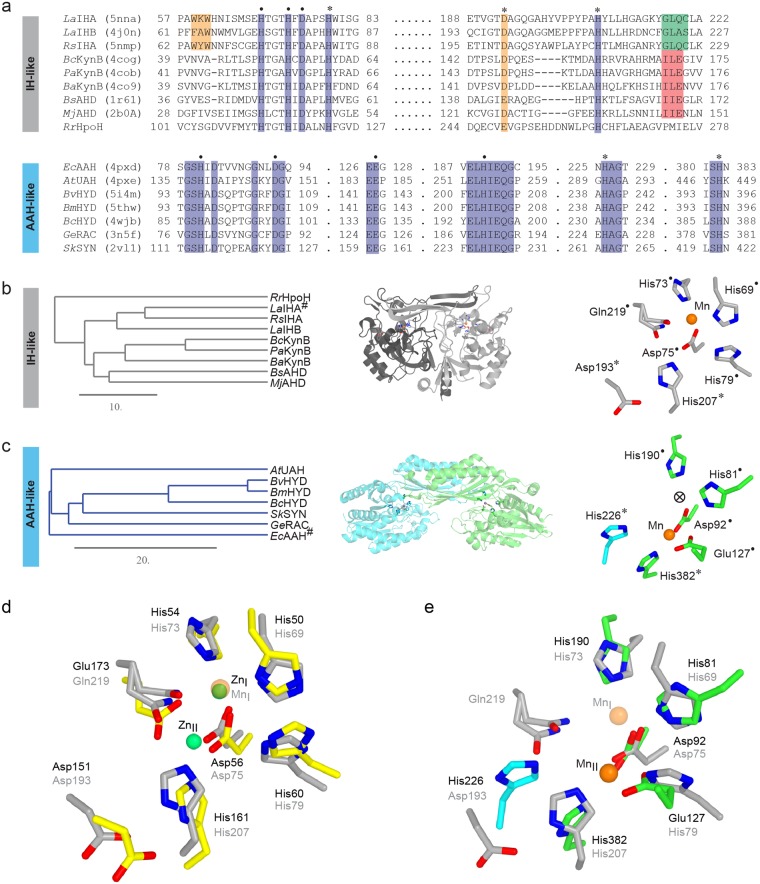
Figure 2.
(a) Alignment of selected sequences of IHA and AAH homologues with known structures. Conserved residues are highlighted in blue. Residues involved in metal coordination are indicated by a dot, residues presumed to be involved in proton transfer are indicated with an asterisk. All are completely conserved, apart from the LaIHA Asp193 (yellow) which is functionally conserved. The orthologue is defining aromatic binding pocket motif for isatin, WXW/FXW, is highlighted in orange. The key GLQC/GLAS motif of IHA and IHB, indicating isatin hydrolase activity, is highlighted in green, with a similarly conserved motif in KynB homologues highlighted in red. (b) Phylogram of IHA homologues from A indicates IHA-like and KynB-like sequences, RrHpoH proposed to carry a novel cyclase activity seems to belong to neither of these. An example of the fold (dark and light grey) and active site position is shown. (c) Phylogram of AAH homologues. Note the large diversity between EcAHH (prokaryote) and AtAAH (eukaryote). An example of the fold (green and cyan) and active site position is shown, His226 (AtAAH) is delivered from opposing monomer through domain swap of AtAAH. (d) Active site residues of BaKynB (Yellow, black numbers) superposed with LaIHA (Grey, grey numbers), the mononuclear Mn2+ in LaIHA is labelled (MnI) and the binuclear Zn2+ sites in BaKynB are labelled ZnI and ZnII. (e) Active site residues of EcAAH (green and cyan, black numbers) superposed manually with LaIHA (grey, grey numbers), the mononuclear Mn2+ from EcAAH labelled MnII.