Abstract
An analysis of 186 diagnostic reports collected from a veterinary clinic in Singapore between 2014 to 2016 showed that sick companion animals can carry bacteria that are of significance to human health. Among the 186 specimens submitted, 82 showed polymicrobial growth (45%, 82/186) and in total, 359 bacteria were isolated. Of the 359 bacteria reported, 45% (162/359) were multi-drug resistant and 18% (66/359) were extended-spectrum-beta-lactamase species. Resistance to broad-spectrum antibiotics were also observed among individual species. Namely, methicillin-resistance among Staphylococcus pseudintermedius (63%, 32/51) and Staphylococcus aureus (50%, 4/8); fluoroquinolone-resistance among Escherichia coli (40%, 17/42) and carbapenem-resistance among Klebsiella pneumoniae (7%, 2/30) were noted. Our analysis suggests that sick pets may contribute to the pool of clinically relevant antibiotic-resistant bacteria and play a role in the spread of antibiotic resistance in Singapore. A more extensive study to better understand the extent of distribution and the factors affecting transmission of antibiotic-resistant bacteria to and from pets is necessary.
Keywords: Pets, Companion animals, Antimicrobial resistance, Antibiotic-resistant bacteria
To the editor
The development of antimicrobial resistance (AMR) in organisms common to food and companion animals (e.g. Escherichia coli, Staphylococcus sp., Enterococcus sp.) and among opportunistic pathogens (e.g. Klebsiella sp., Acinetobacter sp.) has been reported in several countries [1–3]. The transfer of antimicrobial-resistant bacteria from animals to humans is a pressing public health concern particularly when resistance is towards antibiotics used in human medicine [4].
We report a preliminary descriptive analysis investigating the presence of AMR in bacteria isolated from sick companion animals in Singapore. This analysis shall guide further efforts to better understand the contribution of pets to the pool of antibiotic-resistant organisms in the environment and community in Singapore.
One hundred eighty-six (186) veterinary diagnostic reports from a single, community-based private companion animal practice in Singapore, were retrospectively examined. Diagnostic reports, dated 2014 to 2016, contained bacteria isolation results and associated antibiograms of consecutive specimen submissions from pets presenting to the clinic. Bacteria and antimicrobial resistance screening tests were originally requested by veterinarians to guide diagnostic decisions when initial treatment of pathologies was ineffective. Specimens screened were from canine and feline species, and were collected from wounds/abscess (25%, 45/183); ear swabs (24%, 43/183); urine (20%, 37/183) and other samples such as skin and nasal swabs (32%, 58/183). Among these specimens, polymicrobial growth was observed in 53% (24/45) of wound/abscess, 58% (25/43) of ear swab, 22% (8/37) of urine, and 43% (25/58) of all other samples. While there were differences in testing panels, bacteria were screened for susceptibility to antibiotics belonging to at least four of the following antibiotic classes: aminoglycoside, carbapenem, carbolic acid, macrolide, cephalosporin, chloramphenicol, fluoroquinolone, penicillin, rifampin, sulfonamide and tetracycline. Interpretations of antimicrobial susceptibility were based on standard minimum inhibitory concentrations, as described by the Clinical and Laboratory Standards Institute [5].
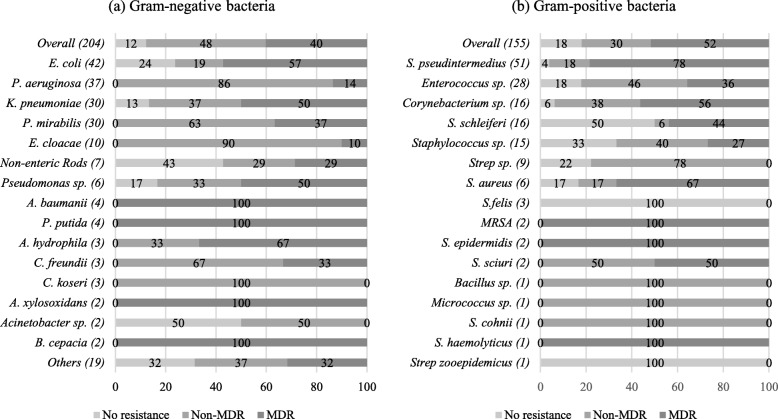
Resistance to multiple antibiotic classes and to broad-spectrum antibiotics was observed among bacteria isolated from sick pets. Forty-five percent (45%, 162/359) of bacteria isolated were multi-drug resistant (MDR), i.e. resistant to at least 3 antibiotic classes [6]; 40% (144/359) were resistant to 1–2 antibiotic classes and 15% (54/359) conferred no resistance to the antibiotics tested. The antimicrobial resistance profiles obtained are illustrated in Fig. 1. Seventy-eight percent (78%, 40/51) of Staphylococcus pseudintermedius, the most frequently detected bacteria (14%, 51/359), were MDR. Fifty-seven percent (57%, 24/42) of E. coli were MDR, and resistance to at least one fluoroquinolone antibiotic (ciprofloxacin, enrofloxacin, marbofloxacin, norfloxacin, ofloxacin) was observed for 40% (17/42) of them. A 50% (15/30) MDR prevalence rate was observed among Klebsiella pneumoniae, and 7% (2/30) showed resistance to carbapenem (imipenem). Multi-drug resistance was also noted in 44% (7/17) of Staphylococcus schleiferi (44%, 7/16) and in 56% (9/16) of Corynebacterium sp.. Among Gram-negative bacteria detected, 32% (66/204) were extended-spectrum-beta-lactamase (ESBL) organisms, as indicated by resistance to 1 or more 3rd-generation cephalosporins (cefotaxime, cefpodoxime, ceftazidime, ceftiofur or ceftizoxime). Forty-seven percent (47%, 14/30) of K. pneumoniae and 40% (17/42) of E. coli were ESBLs. Methicillin-resistance was observed among Gram-positive bacteria, particularly in Staphylococcus spp.. Based on resistance to oxacillin, 50% (4/8) of S. aureus and 63% (32/51) of S. pseudintermedius isolated from sick pets were methicillin-resistant.
Fig. 1.
Antimicrobial resistance profiles of a gram-negative and b gram-positive bacteria isolated from sick pets. Numbers inside brackets ‘()’ indicate number of isolates detected; those on bars indicate percentage per organism; MDR- multi-drug resistant
Our findings show that bacteria of public health relevance – MDR species, ESBLs, methicillin-resistant S. pseudintermedius (MRSP) and S. aureus (MRSA) – could be found among sick pets. The detection of these clinically-relevant bacteria suggests that sick pets, as they live in close proximity with their owners, may play a role in the transmission of AMR between animal and human sources. Conversely, pet owners may also play a role in this transmission. Antimicrobial resistance in bacteria may not necessarily indicate increased pathogenicity, but the presence of MDR organisms in pets, or in any other sources, may affect the resistome and increase the chances of AMR spreading to other bacteria, through lateral gene transfer. Several studies have suggested that commensals, such as the Corynebacterium sp. noted in this analysis, may harbor resistance genes [1, 2, 7] and transfer antimicrobial resistant determinants to opportunistic pathogens [8]. Added measures, such as tighter restrictions to access higher-generation antibiotics, and educational efforts to increase awareness on the prudent use of such antibiotics, may be needed to curtail the spread of AMR in Singapore. Indeed, serious therapeutic challenges may arise if human infections occur due to bacteria that are able to resist the inhibiting effects of broad-spectrum antibiotics (i.e. ESBLs, fluoroquinolone-resistant E. coli and carbapenem-resistant K. pneumoniae). Carbapenem resistance in particular, suggests resistance to nearly all beta-lactam antibiotics commonly used for clinical treatment of gram-negative bacteria [9, 10]. In addition, the transmission of methicillin-resistant S. pseudintermedius to humans, albeit rare, has been increasingly reported by studies citing contact with pets as the likely source of infection [11–13]; pets have likewise been reported to become colonized by MRSA through human contact, then becoming a possible source of re-infection or recurrent MRSA infection for humans [14]. The detection of MDR S. schleiferi was also notable as this bacteria has been reported to cause both veterinary and human skin infections [15].
This initial assessment is confined to data obtained from retrospective veterinary diagnostic reports. These reports did not indicate whether the isolated bacteria were the causative agents of infection. Information on specimens such as faeces, that are not commonly sent for antibiotic resistance screening, were unavailable. Data on the pet’s prior antibiotic use, veterinary care or interaction with human carriers (if any) that may have increased chances of exposure to antibiotic-resistant bacteria were also unavailable from the diagnostic reports. Nevertheless, our analysis suggests that sick pets may contribute to the pool of clinically relevant antibiotic-resistant bacteria in Singapore and that a more extensive epidemiological study on AMR bacteria carried by sick pets is necessary to better understand the extent of distribution, the direction and risk factors pertaining to transmission as well as the associations between veterinary, environmental and clinical AMR data.
Acknowledgments
We thank the National Environment Agency for operational support; Ms. Jean Lee Ryan of Companion Animal Surgery Pte Ltd. (Singapore) for providing the diagnostic reports used as the data source for this study and Dr. Shawn Ting of the Agri-Food and Veterinary Authority for comments on the paper. We also thank Dr. Ng Lee Ching of the Environmental Health Institute, National Environment Agency for providing scientific guidance and support.
Funding
This study was funded by the National Environment Agency-Singapore.
Availability of data and materials
Please contact corresponding author for data requests.
Abbreviations
- AMR
Antimicrobial resistance
- ESBL
Extended-spectrum-beta-lactamase
- MDR
Multi-drug resistant
- MRSA
Methicillin-resistant Staphylococcus aureus
- MRSP
Methicillin-resistant Staphylococcus pseudintermedius
Authors’ contributions
SHPH, CML, LF and RAG were involved in the conception and design of the study. SHPH, LF and RAG facilitated the transfer of diagnostic reports from the veterinary clinic to the research institute. SHPH and AZBMA collated data from veterinary reports; SHPH, CML and RAG were involved in the analysis and interpretation of the data. KTA and JKSL provided technical input regarding antimicrobial resistance. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sri Harminda Pahm Hartantyo, Email: Sri_Harminda_HARTANTYO@nea.gov.sg.
Man Ling Chau, Email: CHAU_Man_Ling@nea.gov.sg.
Laurent Fillon, Email: fillonlaurent@hotmail.fr.
Ahmad Zhafir Bin Mohamad Ariff, Email: Ahmad_Zhafir_ARIFF@nea.gov.sg.
Joanne Su Lin Kang, Email: Joanne_KANG@nea.gov.sg.
Kyaw Thu Aung, Email: AUNG_Kyaw_Thu@nea.gov.sg.
Ramona Alikiiteaga Gutiérrez, Phone: +65 6571 0476, Email: Ramona_GUTIERREZ@nea.gov.sg.
References
- 1.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall BM, Ochieng DJ, Levy SB. Commensals: underappreciated reservoir of antibiotic resistance. Microbe. 2009;4(5):231–238. [Google Scholar]
- 3.Murphy CP, Reid-Smith RJ, Boerlin P, Weese JS, Prescott JF, Janecko N, Hassard L, McEwan SA. Escherichia coli and selected veterinary and zoonotic pathogens isolated from environmental sites in companion animal veterinary hospitals in southern Ontario. Can Vet J. 2010;51(9):963–972. [PMC free article] [PubMed] [Google Scholar]
- 4.Guardabassi L, Schwarz S, Lloyd DH. Pet animals as reservoirs of antimicrobial-resistant bacteria. J Antimicrob Chemother. 2004;54:321–332. doi: 10.1093/jac/dkh332. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and laboratory standards institute (CLSI): performance standards for antimicrobial susceptibility testing. In: Twenty-Fourth Informational Supplement M100-S24. Wayne: CLSI; 2014.
- 6.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2011;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 7.Andremont A. Commensal flora may play key role in spreading antibiotic resistance. ASM News. 2003;69(12):601–7.
- 8.Schmidt VM, Williams NJ, Pinchbeck G, Corless CE, Shaw S, McEwan N, Dawson S, Nuttall T. Antimicrobial resistance and characterisation of staphylococci isolated from healthy Labrador retrievers in the United Kingdom. BMC Vet Res. 2014;10(17):1–14. doi: 10.1186/1746-6148-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitout JDD, Laupland KB. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8(3):159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 10.Woodford N, Wareham DW, Guerra B, Teale C. Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: an emerging public health risk of our own making? J Antimicrob Chemother. 2014;69(2):287–291. doi: 10.1093/jac/dkt392. [DOI] [PubMed] [Google Scholar]
- 11.Duijkeren Ev CB, Greko C, Moreno MA, Pomba MC, Pyorala S, Ruzuskas M, Sanders P, Threlfall EJ, Torren-Edo J, et al. Review on methicillin-resistant Staphylococcus pseudintermedius. J Antimicrob Chemother. 2011;66:2705–2714. doi: 10.1093/jac/dkr367. [DOI] [PubMed] [Google Scholar]
- 12.Somayaji R, Priyantha MA, Rubin JE, Church D. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: 24 cases. Diagn Microbiol Infect Dis. 2016;85(4):471–476. doi: 10.1016/j.diagmicrobio.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Stegmann R, Burnens A, Maranta CA, Perreten V. Human infection associated with methicillin-resistant Staphylococcus pseudintermedius ST71. J Antimicrob Chemother. 2010;65(9):2047–2048. doi: 10.1093/jac/dkq241. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira JP, Anderson KL, Correa MT, Lyman R, Ruffin F, Reller B, Fowler VG. Transmission of MRSA between companion animals and infected human patients presenting to outpatient medical care facilities. PLoS One. 2011;6(11):1–6. doi: 10.1371/journal.pone.0026978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis MF, Cain C, Brazil AM, Rankin SC. Two coagulase-negative staphylococci emerging as potential zoonotic pathogens: wolves in sheep’s clothing? Front Microbiol. 2013;3(123):1–4. doi: 10.3389/fmicb.2013.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Please contact corresponding author for data requests.