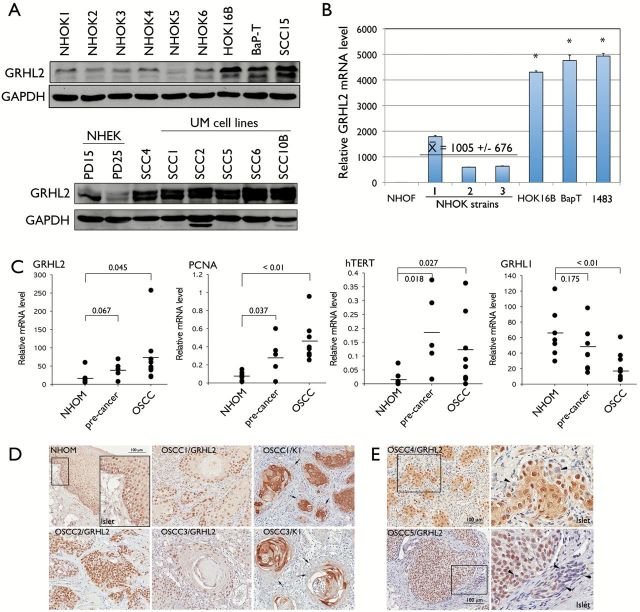
Figure 1.
GRHL2 expression is enhanced in cells and tissues of OSCCs. (A) Western blotting was performed for GRHL2 using whole cell extracts from six different strains of NHOK, immortalized cells (HOK-16B) and OSCC cell lines (HOK-16B/BaP-T and SCC15). Another blot containing whole cell extracts from NHEK and OSCC cell lines, including SCC4 and those from UM-OSCCs were probed for GRHL2 protein levels. GAPDH was used as loading control. PD refers to population doublings. (B) qRT-PCR was performed with RNA extracted from NHOF, three strains of rapidly proliferating NHOK and the established cell lines (HOK-16B, HOK-16B/BaP-T and 1483). Bars indicate standard deviation and the asterisk (*) indicates statistical significance (P < 0.05), compared with the mean values of the control group (NHOK). (C) Epithelial tissues were excised from NHOM, preneoplastic oral lesions and OSCC tissues by laser-captured microdissection (LCM). mRNA levels of GRHL2, PCNA, hTERT and GRHL1 were determined from each sample by qRT-PCR. P values are shown in each group. (D) IHC was performed with NHOM, preneoplastic oral lesions and OSCC tissues using α-GRHL2 or α-K1 antibody. Representative staining results were shown for various OSCC samples (OSCC1–OSCC5). GRHL2 and K1 staining patterns appear to be mutually exclusive; tumor cells with strong GRHL2 appear to be completely lacking K1 staining (arrows). (E) In some tumors, GRHL2 expression pattern was found heterogeneous within the same tumor island; some cells exhibited strong GHRL2 staining while others completely lacked (arrowheads). Right-side panel shows higher magnification of the view field within the boxed area.