Abstract
Saccharomyces sensu stricto complex consist of yeast species, which are not only important in the fermentation industry but are also model systems for genomic and ecological analysis. Here, we present the complete genome assemblies of Saccharomyces jurei, a newly discovered Saccharomyces sensu stricto species from high altitude oaks. Phylogenetic and phenotypic analysis revealed that S. jurei is more closely related to S. mikatae, than S. cerevisiae, and S. paradoxus. The karyotype of S. jurei presents two reciprocal chromosomal translocations between chromosome VI/VII and I/XIII when compared to the S. cerevisiae genome. Interestingly, while the rearrangement I/XIII is unique to S. jurei, the other is in common with S. mikatae strain IFO1815, suggesting shared evolutionary history of this species after the split between S. cerevisiae and S. mikatae. The number of Ty elements differed in the new species, with a higher number of Ty elements present in S. jurei than in S. cerevisiae. Phenotypically, the S. jurei strain NCYC 3962 has relatively higher fitness than the other strain NCYC 3947T under most of the environmental stress conditions tested and showed remarkably increased fitness in higher concentration of acetic acid compared to the other sensu stricto species. Both strains were found to be better adapted to lower temperatures compared to S. cerevisiae.
Keywords: evolution, fitness, PacBio, translocation, Saccharomyces
Saccharomyces sensu stricto yeasts, currently comprise eight species: S. cerevisiae, S. paradoxus, S. uvarum, S. mikatae, S. kudriavzevii, S. arboricola, S. eubayanus, S. jurei (Martini and Martini 1987; Wang and Bai 2008; Naumov et al. 2000; Naumov et al. 1995a; Naumov et al. 1995b; Libkind et al. 2011; Naseeb et al. 2017b) and two natural hybrids: S. pastorianus (Masneuf et al. 1998; Querol and Bond 2009) and S. bayanus (Nguyen et al. 2011). Saccharomyces jurei is the latest addition to the sensu stricto clade and was isolated from oak tree bark and surrounding soil at an altitude of 1000 m above sea level in Saint Auban, France (Naseeb et al. 2017b). It is known that species within the sensu stricto group are reproductively isolated and possess post- zygotic barriers (Naumov 1987). Moreover, yeasts within this group exhibit almost identical karyotypes with 16 chromosomes (Cardinali and Martini 1994; Carle and Olson 1985; Naumov et al. 1996).
In the modern era of yeast genetics, the advances in sequencing technology have lead to the whole genome sequencing of many Saccharomyces sensu stricto species (S. cerevisiae, S. bayanus var. uvarum, S. kudriavzevii, S. mikatae, S. paradoxus, S. eubayanus and S. arboricola) (Libkind et al. 2011; Liti et al. 2013; Cliften et al. 2003; Kellis et al. 2003; Scannell et al. 2011; Casaregola et al. 2000). To date, more than 1000 S. cerevisiae strains belonging to different geographical and environmental origins have been sequenced and assembled (Engel and Cherry 2013; Peter et al. 2018). The availability of sequencing data from multiple strains of Saccharomycotina yeast species has enhanced our understanding of biological mechanisms and comparative genomics. Researchers are now combining comparative genomics with population ecology to better understand the genetic variations, taxonomy, evolution and speciation of yeast strains in nature. Genome variation provides the raw material for evolution, and may arise by various mechanisms including gene duplication, horizontal gene transfer, hybridization and micro and macro rearrangements (Fischer et al. 2001; Seoighe et al. 2000; Lynch 2002; Hall et al. 2005; Naseeb et al. 2017a; Naseeb et al. 2016; Naseeb and Delneri 2012). Synteny conservation studies have shown highly variable rates of genetic rearrangements between individual lineages both in vertebrates and in yeasts (Bourque et al. 2005; Fischer et al. 2006; Vakirlis et al. 2016). This genome variation is a means of evolutionary adaptation to environmental changes. An understanding of the genetic machinery linked to phenotypic variation provides knowledge of the distribution of Saccharomyces species in different environments, and their ability to withstand specific conditions (Goddard and Greig 2015; Jouhten et al. 2016; Brice et al. 2018; Peter et al. 2018).
Recently, we isolated two strains (NCYC 3947T and NCYC 3962) of Saccharomyces jurei from Quercus robur bark and surrounding soil (Naseeb et al. 2017b). The initial sequencing of ITS1, D1/D2 and seven other nuclear genes showed that both strains of S. jurei were closely related to S. mikatae and S. paradoxus and grouped in Saccharomyces sensu stricto complex. We also showed that S. jurei can readily hybridize with other sensu stricto species but the resulting hybrids were sterile (Naseeb et al. 2017b). Here, we represent high quality de novo sequence and assembly of both strains (NCYC 3947T and NCYC 3962) of S. jurei. The phylogenetic analysis placed S. jurei in the sensu stricto clade, in a small monophyletic group with S. mikatae. By combining Illumina HiSeq and PacBio data, we were able to assemble full chromosomes and carry out synteny analysis. Moreover, we show that S. jurei NCYC 3962 had higher fitness compared to NCYC 3947T under different environmental conditions. Fitness of S. jurei strains at different temperatures showed that it was able to grow at wider range of temperatures (12°-37°).
Material and Methods
Yeast strains
Strains used in this study are presented in Table 1. All strains were grown and maintained on YPDA (1% w/v yeast extract, 2% w/v Bacto-peptone, 2% v/v glucose and 2% w/v agar). Species names and strains number are stated in Table 1.
Table 1. Strains used in this study.
| Species | Strain number | References |
|---|---|---|
| S. jurei | NCYC 3947T | (Naseeb et al. 2017b) |
| NCYC 3962 | ||
| S. cerevisiae | NCYC 505T | (Vaughan Martini and Kurtzman 1985) |
| S. paradoxus | CBS 432T | (Naumov 1987) |
| S. mikatae | NCYC 2888T (IFO 1815T) | (Yamada et al. 1993) |
| S. kudriavzevii | NCYC 2889T (IFO 1802T) | (Yamada et al. 1993) |
| S. arboricola | CBS 10644T | (Wang and Bai 2008) |
| S. eubayanus | PYCC 6148T (CBS 12357T) | (Libkind et al. 2011) |
| S. uvarum | NCYC 2669 (CBS 7001) | (Pulvirenti et al. 2000) |
| S. pastorianus | NCYC 329T (CBS 1538T) | (Martini and Martini 1987) |
DNA Extraction
For Illumina Hiseq, the total DNA was extracted from an overnight grown culture of yeast strains by using the standard phenol/chloroform method described previously (Fujita and Hashimoto 2000) with some modifications. Briefly, 5 ml of overnight grown yeast cells were centrifuged and resuspended in 500 μl EB buffer (4M sorbitol, 500mM EDTA and1M DTT) containing 1 mg/ml lyticase. The cells were incubated at 37° for 1 hr. Following incubation, the cells were mixed with stop solution (3M NaCl, 100mM Tris pH 7.5 and 20mM EDTA) and 60 μl of 10% SDS. The cell suspension was vortexed and mixed with 500 μl phenol-chloroform. The samples were centrifuged at 13000 rpm for 2 min to separate the aqueous phase from the organic phase. The upper aqueous phase was transferred to a clean 1.5 ml tube and phenol-chloroform step was repeated twice until a white interface was no longer present. The aqueous phase was washed with 1 ml absolute ethanol by centrifugation at 13000 rpm for 10 min. The pellet was air dried and resuspended in 30 μl of sterile milliQ water.
Genomic DNA for PacBio sequencing was extracted using Qiagen Genomic-tip 20/G kit (cat. No. 10223) following manufacturer’s recommended instructions. The yield of all DNA samples was assessed by the nanodrop spectrophotometer (ND-1000) and by Qubit 2.0 fluorometer (catalog no. Q32866). Purity and integrity of DNA was checked by electrophoresis on 0.8% (w/v) agarose gel and by calculating the A260/A280 ratios.
Library preparation for Illumina and PacBio sequencing
Paired end whole-genome sequencing was performed using the Illumina HiSeq platform. FastQC (Babraham Bioinformatics) was used to apply quality control to sequence reads, alignment of the reads was performed using BOWTIE2 (Langmead and Salzberg 2012) and post-processed using SAMTOOLS (Li et al. 2009).
For Pacbio sequencing, genomic DNA (10 μg) of NCYC 3947T and NCYC 3962 strains was first DNA damage repaired, sheared with Covaris G-tube, end repaired and exonuclease treated. SMRTbell library (10-20kb size) was prepared by ligation of hairpin adaptors at both ends according to PacBio recommended procedure (Pacific Bioscience, No: 100-259-100). The resulting library was then size selected using Blue Pippin with 7-10kb cut-off. Sequencing run was performed on PacBio RS II using P6/C4 chemistry for 4 hr. The genome was assembled using SMRT analysis and HGAP3 pipeline was made using default settings.
Genome assembly, annotation, orthology and chromosomal structural plots
The PacBio sequences were assembled using hierarchical genome-assembly process (HGAP) (Chin et al. 2013). Protein coding gene models were predicted using Augustus (Stanke and Morgenstern 2005) and the Yeast Genome Annotation Pipeline (Byrne and Wolfe 2005). In addition, protein sequences from other Saccharomyces species were aligned to the genome assembly using tblastn (Gertz et al. 2006). These predictions and alignments were used to produce a final set of annotated genes with the Apollo annotation tool (Lewis et al. 2002). The protein sequences were functionally annotated using InterproScan (Jones et al. 2014). Orthologous relationships with S. cerevisiae S288C sequences were calculated using InParanoid (Berglund et al. 2008). Non-coding RNAs were annotated by searching the RFAM database (Nawrocki et al. 2015) using Infernal (Nawrocki and Eddy 2013). Further tRNA predictions were produced using tRNAscan (Lowe and Eddy 1997). Repeat sequences were identified in Repbase (Bao et al. 2015) using Repeat Masker (Smit et al. 2013–2015). The dotplots were constructed by aligning S. jurei genome to the S. cerevisiae S288C genome using NUCmer and plotted using MUMmerplot (Kurtz et al. 2004). These features are available to browse via a UCSC genome browser (Kent et al. 2002) track hub (Raney et al. 2014). Single nucleotide polymorphisms (SNPs) were identified using Atlas-SNP2(Challis et al. 2012).
Phenotypic assays
Temperature tolerance:
Fitness of S. jurei strains and Saccharomyces sensu stricto type strains was examined using FLUOstar optima microplate reader at 12°, 16°, 20°, 25°, 30° and 37°. Cells were grown from a starting optical density (OD) of 0.15 to stationary phase in YPD (1% w/v yeast extract, 2% w/v Bacto-peptone and 2% w/v glucose) medium. The growth OD595 was measured every 5 min with 1 min shaking for 72 hr. Growth parameters, lag phase (λ), maximum growth rate (µmax), and maximum biomass (Amax) were estimated using R shiny app on growth curve analysis (https://kobchai-shinyapps01.shinyapps.io/growth_curve_analysis/).
Environmental stress:
Strains were screened for tolerance to environmental stressors using a high-throughput spot assay method. Cells were grown in a 96-well plate containing 100 µl YPD in four replicates at 30° for 48 hr. The yeast strains grown in 96-well plate were sub-cultured to a 384 well plate to achieve 16 replicates of each strain and grown at 30° for 48 hr. Singer ROTOR HDA robot (Singer Instruments, UK) was used to spot the strains on (i) YPDA + 0.4% & 0.6% acetic acid, (ii) YPDA+ 4mM & 6mM H2O2, (iii) YPDA+ 2.5mM & 5mM CuSO4, (iv) YPDA+ 2% & 5% NaCl, (v) YPDA+ 5% & 10% Ethanol (vi) YPA+ 15% maltose and (vii) YPA+ 30% & 35% glucose. The spot assay plates were incubated at 30° and high-resolution images of phenotypic plates were taken using phenobooth after 3 days of incubation (Singer Instruments, UK). The colony sizes were calculated in pixels using phenosuite software (Singer Instruments, UK) and the heat maps of the phenotypic behaviors were constructed using R shiny app (https://kobchai-shinyapps01.shinyapps.io/heatmap_construction/).
Data and reagent availability
Strains are available upon request. Supplemental files are available at FigShare (https://figshare.com/s/60bbbc1e98886077182a). Figure S1 shows alignment of the amino acid sequences of MEL1 gene belonging to S. jurei NCYC 3947T (Sj) and S. mikatae IFO 1816 (Sm). Table S1, Table S2, Table S3 and Table S4 list the genes, which are present in simple one to one orthologous relationship, in many to many relationship, in many to one relationship and in one to many relationship, respectively. Table S5 lists the genes that are present in S. cerevisiae but absent in S. jurei. Table S6 lists the genes which are present in S. jurei but absent in S. cerevisiae. Table S7 lists the genes which are used to construct the phylogenetic tree. Table S8 lists the genes which are potentially introgressed in S. jurei genome from S. paradoxus. Table S9, Table S10 and Table S11 show lag phase time (λ), maximum growth rate (µmax) and maximum biomass (Amax) of Saccharomyces species used in this study, respectively. The sequences and annotations reported in this paper are available in the European Nucleotide Archive under project ID PRJEB24816, assembly ID GCA_900290405 and accession number ERZ491603.
Results and Discussion
High quality de novo sequencing and assembly of S. jurei genome
Genome sequencing of the diploid S. jurei NCYC 3947T and NCYC 3962 yeast strains was performed using Illumina Hiseq and Pacbio platforms. We obtained approximately 9.02 × 105 and 4.5 × 105 reads for NCYC 3947T and NCYC 3962 respectively. We obtained 2 × 101 bp reads derived from ∼200 bp paired-end reads which were assembled in 12 Mb genome resulting in a total coverage of 250x based on high quality reads. The sequencing results and assembled contigs are summarized in Tables 2, 3, and 4. By combining the Illumina mate pair and Pacbio sequencing we were able to assemble full chromosomes of S. jurei NCYC 3947T and NCYC 3962 (Tables 5 and 6). The total genome size (∼12 Mb) obtained for both strains of S. jurei was comparable to the previously published genomes of Saccharomyces sensu stricto species (Scannell et al. 2011; Goffeau et al. 1996; Liti et al. 2013; Baker et al. 2015).
Table 2. Summary of S. jurei NCYC 3947T genome sequencing and assembly using Hi-seq platform.
| Metric | Contigs | Contigs >= 500bp | Scaffolds | Scaffolds >= 500bp |
|---|---|---|---|---|
| Number | 810 | 250 | 753 | 211 |
| Total Length | 11,938,007 | 11,869,594 | 11,940,421 | 11,869,594 |
| Length Range | 87-673,524 | 525-673,524 | 87-673,524 | 525-673,524 |
| Average Length | 14,738 | 56,254 | 15,857 | 56,254 |
| N50 | 172,207 | 279,631 | 279,631 | 279,631 |
Table 3. Summary of S. jurei NCYC 3962 genome sequencing and assembly using Hi-seq platform.
| Metric | Contigs | Contigs >= 500bp | Scaffolds | Scaffold >= 500bp |
|---|---|---|---|---|
| Number | 3719 | 987 | 3618 | 933 |
| Total length | 11,760,925 | 11,419,281 | 11,768,034 | 11,441,494 |
| Length range | 59-80,684 | 507-80,684 | 59-80,684 | 507-80,684 |
| Average length | 3,162 | 11,569 | 3,252 | 12,263 |
| N50 | 20,806 | 21,318 | 21,928 | 22,552 |
Table 4. Summary of S. jurei NCYC 3947T and NCYC 3962 genome assembly using PacBio platform.
| Metric | S. jurei NCYC 3947 | S. jurei NCYC 3962 |
|---|---|---|
| Contigs | 35 | 57 |
| Max contig length | 1,474,466 | 1,470,125 |
| Contig N50 | 738,741 | 652,030 |
| Total assembly size | 12,306,756 | 12,932,708 |
Table 5. Total lengths of chromosomes assembled in S. jurei NCYC 3947T.
| Sequence name | Length (bp) including gaps |
|---|---|
| chrI.1_chrXIII.2 | 809,572 |
| chrII | 809,280 |
| chrIII | 308,350 |
| chrIV | 1,474,466 |
| chrV | 584,553 |
| chrVI.1_chrVII.2 | 730,011 |
| chrVI.2_chrVII.1 | 638,210 |
| chrVIII | 534,462 |
| chrIX | 434,517 |
| chrX | 738,741 |
| chrXI | 671,067 |
| chrXII.1 | 458,950 |
| chrXII.2 | 568,540 |
| chrI.2_chrXIII.1 | 334,136 |
| chrXIV | 749,072 |
| chrXV | 1,068,672 |
| chrXVI | 920,427 |
| chrMT | 105,732 |
Table 6. Total lengths of chromosomes assembled in S. jurei NCYC 3962.
| Sequence name | Length (bp) including gaps |
|---|---|
| chrI.1_chrXIII.2 | 756,315 |
| chrII | 814,183 |
| chrIII | 329,028 |
| chrIV | 1,470,125 |
| chrV | 570,437 |
| chrVI.1_chrVII.2 | 723,619 |
| chrVII.2_chrVI.1 | 652,030 |
| chrVIII | 536,516 |
| chrIX | 439,662 |
| chrX.1 | 487,336 |
| chrX.2 | 258,684 |
| chrXI | 676,065 |
| chrXII.1 | 475,978 |
| chrXII.2 | 571,082 |
| chrI.2_chrXIII.1 | 334,998 |
| chrXIV | 790,124 |
| chrXV.1 | 474,048 |
| chrXV.2 | 240,703 |
| chrXV.3 | 236,823 |
| chrXV.4 | 114,889 |
| chrXVI | 806,586 |
| chrMT | 110,829 |
S. jurei genome prediction and annotation
The high-quality de novo assembly of S. jurei NCYC 3947T genome resulted in 5,794 predicted protein-coding genes for S. jurei, which is similar to the published genomes of other sensu stricto species (Baker et al. 2015; Liti et al. 2009; Liti et al. 2013; Scannell et al. 2011; Walther et al. 2014). Of the predicted protein-coding genes, 5,124 were in a simple 1:1 putatively orthologous relationship between S. cerevisiae and S. jurei (Table S1). From the remaining protein-coding genes, 35 genes showed many to many relationship (multiple S. cerevisiae genes in paralogous cluster with multiple S. jurei genes (Table S2), 31 genes were in many to one relationship (many genes in S. cerevisiae are in an paralogous cluster with a single S. jurei gene; most of these were found to be retrotransposons; Table S3) and 50 genes were in one to many relationships (one S. cerevisiae gene in an paralogous cluster with many S. jurei genes; Table S4). Interestingly, we found an increase in the copy number of maltose metabolism and transport genes (IMA1, IMA5, MAL31, and YPR196W- 2 copies of each gene), flocculation related gene (FLO1- 2 copies) and hexose transporter (HXT8- 3 copies). Increased dosage of these genes in S. jurei could have conferred selective advantage toward better sugar utilization (Lin and Li 2011; Ozcan and Johnston 1999; Soares 2011; Adamczyk et al. 2016). Genes encoding for PAU proteins (a member of the seripauperin multigene family), copper resistance and salt tolerance related genes were found to be present in fewer copies in S. jurei genome compared to S. cerevisiae. This variation in copy number of genes in a genome can have phenotypic and physiological effects on the species (Landry et al. 2006; Adamo et al. 2012; Gorter de Vries et al. 2017).
We also searched for the presence of repetitive elements in S. jurei NCYC 3947T and NCYC 3962 using BLAST and compared them to the Ty elements in S. cerevisiae. We detected Ty1-LTR, Ty2-LTR, Ty2-I-int, Ty3-LTR, Ty3-I and Ty4 sequences in both strains of S. jurei. Interestingly, we found an increased number of Ty1-LTR, Ty2-LTR, Ty3-LTR and Ty4 elements in S. jurei genome compared to S. cerevisiae (Table 7). High copy numbers of Ty1, Ty2, and Ty3 transposable elements have also been reported in different strains of S. cerevisiae, e.g., Ty1 and Ty2 in French Dairy, Ty2 in Alpechin, Ty1 in Mexican Agave, and Ty3 in Ecuadorean clade (Peter et al. 2018; Bleykasten-Grosshans et al. 2013). Repetitive sequences are found in genomes of all eukaryotes and can be a potential source of genomic instability since they can recombine and cause chromosomal rearrangements, such as translocations, inversions and deletions (Naseeb et al. 2016; Shibata et al. 2009; Chan and Kolodner 2011).
Table 7. Counts of Ty elements in S. cerevisiae, S. jurei NCYC 3947T and NCYC 3962.
| Ty elements | Ty elements annotation | Counts in S. cerevisiae | Counts in S. jurei NCYC 3947T | Counts in S. jurei NCYC 3962 |
|---|---|---|---|---|
| Ty | Yeast Ty transposable element Ty-pY109 near tRNA-Lys1 gene | 164 | 71 | 74 |
| Ty1-LTR | Ty1 LTR-retrotransposon from yeast (LTR) | 124 | 276 | 272 |
| Ty2-LTR | Ty2 LTR-retrotransposon from yeast (LTR) | 108 | 118 | 117 |
| Ty2-I-int | Ty2 LTR-retrotransposon from yeast (internal portion). | 15 | 2 | 2 |
| Ty3-LTR | S. paradoxus Ty3-like retrotransposon, Long terminal repeat | 61 | 70 | 71 |
| Ty3-I | S. paradoxus Ty3-like retrotransposon, Internal region. | 2 | 1 | 1 |
| Ty4 | Gag homolog, Ty4B = protease, integrase, reverse transcriptase,and RNase H domain containing protein {retrotransposon Ty4} | 51 | 164 | 162 |
Saccharomyces jurei share a chromosomal translocation With Saccharomyces mikatae IFO 1815
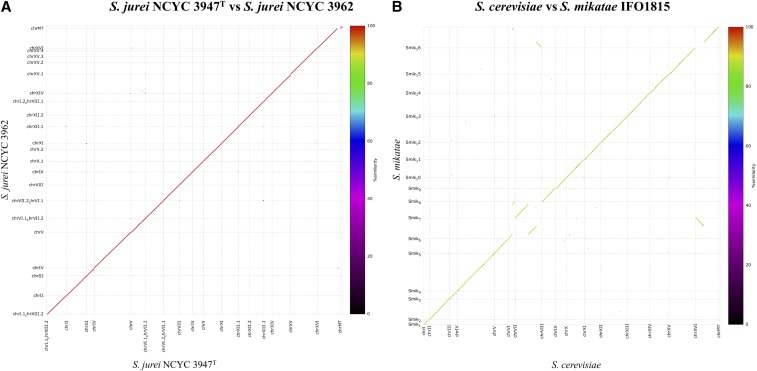
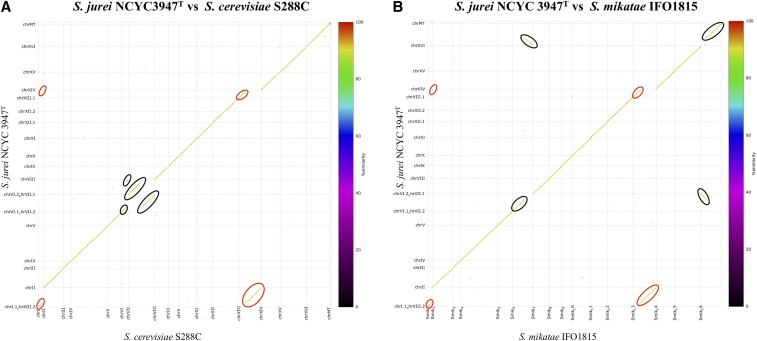
To check the presence or absence of genomic rearrangements in S. jurei, we compared the chromosome structures between S. jurei NCYC 3947T and S. jurei NCYC 3962 (Figure 1A), between S. cerevisiae S288C and S. mikatae IFO1815 (Figure 1B), between S. jurei NCYC 3947T and S. cerevisiae S288C (Figure 2A) and between S. jurei NCYC 3947T and S. mikatae IFO1815 (Figure 2B). The two S. jurei strains had a syntenic genome (Figure 1A), while we identified two chromosomal translocations with S. cerevisiae S288C (Figure 2A). One translocation is unique to S. jurei and is located between chromosomes I and XIII (Figure 2, red ovals), while the second translocation is located between chromosomes VI and VII in the same position of the previously identified translocation in S. mikatae IFO1815 (Figure 2, black ovals).
Figure 1.
Dot plot alignments comparing the chromosome sequence identity of S. jurei NCYC 3947T vs. S. jurei NCYC 3962 (A) and S. cerevisiae S288C vs. S. mikatae IFO1815 (B). The broken lines represent chromosomal translocations between chromosomes VI / VII and XVI / VII.
Figure 2.
Dot plot alignments comparing the chromosome sequence identity of S. jurei NCYC 3947T vs. S. cerevisiae S288C (A) and S. jurei NCYC 3947T vs. S. mikatae IFO1815 (B). Black ovals represent the translocation between chromosomes VI and VII, which is common in S. mikatae and S. jurei whereas red ovals represent the translocation between chromosomes I and XIII, which is unique to S. jurei.
The breakpoints of the translocation I/XIII are in the intergenic regions between uncharacterized genes. The breakpoints neighborhood is surrounded by several Ty elements (Ty1-LTR, Ty4, and Ty2-LTR) and one tRNA, which may have caused the rearrangement (Bridier-Nahmias et al. 2015; Fischer et al. 2000; Liti et al. 2013; Mieczkowski et al. 2006). The translocation in common with S. mikatae shares the same breakpoints between open reading frames (ORFs) YFR006W and YFR009W on chromosome VI, and between ORFs YGR021W and YGR026W on chromosome VII. This translocation is also shared by both strains of S. mikatae, but not with other Saccharomyces sensu stricto species. Overall this suggests a common evolutionary history between these strains and species, however an adaptive value of this rearrangement or a case of breakpoint re-usage cannot be ruled out since rearrangements can be adaptive with evidence both from nature and lab setting. (Chang et al. 2013; Dunham et al. 2002; Avelar et al. 2013; Colson et al. 2004; Adams et al. 1992; Fraser et al. 2005; Hewitt et al. 2014). Several natural isolates of S. cerevisiae present karyotypic changes (Hou et al. 2014) and the reciprocal translocation present between chromosomes VIII and XVI is able to confer sulphite resistance to the yeasts strains in vineyards (Perez-Ortin et al. 2002). Furthermore, lab experimental evolution studies in different strains of S. cerevisiae when evolved under similar condition end up sharing the same breakpoints (Dunham et al. 2002). Previous studies on mammalian systems have shown that breakpoints maybe reused throughout evolution at variable rates (Larkin et al. 2009; Murphy et al. 2005), and breakpoint re-usage has also been found between different strains of S. pastorianus (Hewitt et al. 2014).
Novel genes present in S. jurei
The comparison between S. jurei and S. cerevisiae genome showed 622 differentially present genes. 179 open reading frames (ORFs) were predicted to be novel in S. jurei when compared to S. cerevisiae reference S288C strain (Table S5). To further confirm if these ORFs were truly novel, we analyzed the sequences in NCBI nucleotide database and in Saccharomyces Genome Database (SGD) against all the fungal species. We found 4 novel ORFs that have no significant match to any of the available genomes (Table S5-shown in red). 5 ORFs gave partial similarity to different fungal species such as Vanderwaltozyma polyspora, Kluyveromyces marxianus, Torulaspora delbrueckii, Zygosaccharomyces rouxii, Hyphopichia burtonii, Kazachstania africana, Trichocera brevicornis, Lachancea walti, and Naumovozyma castellii (Table S5-yellow highlighted). Majority of the remaining sequences gave full or partial matches to S. cerevisiae natural isolates (Strope et al. 2015; Peter et al. 2018), S. paradoxus, S. mikatae, S. kudriavzevii, S. bayanus, S. uvarum, and S. eubayanus.
Moreover, we also found 462 ORFs, which are present in S. cerevisiae genome but were lost in S. jurei (Table S6). The Gene Ontology (GO) analysis of these genes showed significant enrichment of RNA-directed DNA polymerase activity, aryl-alcohol dehydrogenase (NAD+) activity, DNA-directed DNA polymerase activity, and asparaginase activity. The majority of genes which were novel or lost in S. jurei were found to be subtelomeric or telomeric, in regions known to show higher genetic variations (Bergström et al. 2014).
The genes lost in S. jurei encompass functionally verified ORFs, putative genes and uncharacterized genes. Some of the verified ORFs included ribosomal subunits genes, asparagine catabolism genes, alcohol dehydrogenase genes, hexose transporters, genes involved in providing resistance to arsenic compounds, phosphopyruvate hydratase genes, iron transport facilitators, ferric reductase genes and flocculation related genes.
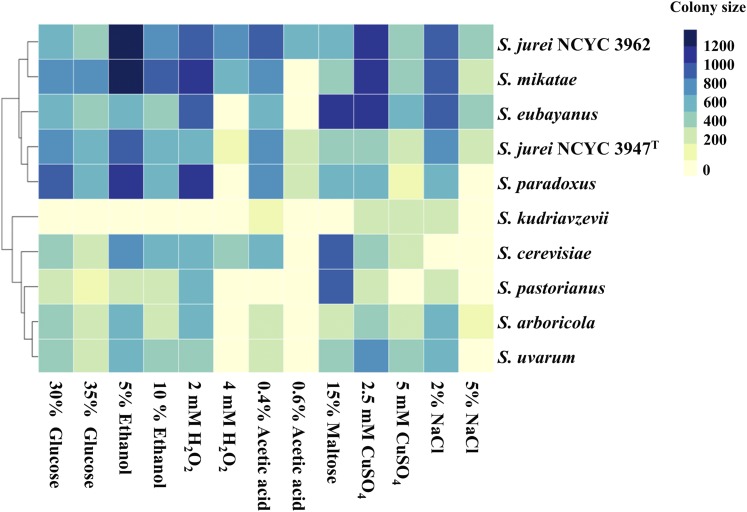
We found that S. jurei genome lacks four out of seven alcohol dehydrogenase (AAD) genes including the functional AAD4 gene, which is involved in oxidative stress response (Delneri et al. 1999a; Delneri et al. 1999b). Although S. jurei has lost AAD4 gene, however, it was able to tolerate oxidative stress caused by 4mM H2O2 (Figure 3A).
Figure 3.
Heat map representing phenotypic fitness of S. jurei NCYC 3947T and NCYC 3962 compared to sensu stricto species type strains in response to different environmental stressors at 30°C. Phenotypes are represented with colony sizes calculated as pixels and colored according to the scale, with light yellow and dark blue colors representing the lowest and highest growth respectively. Hierarchical clustering of the strains is based on the overall growth profile under different media conditions tested.
All four genes of the ASP3 gene cluster located on chromosome XII are absent in S. jurei. It was not surprising since this gene cluster is only known to be present in S. cerevisiae strains isolated from industrial and laboratory environments and lost from 128 diverse fungal species (Gordon et al. 2009; League et al. 2012). These genes are up-regulated during nitrogen starvation allowing the cells to grow by utilizing extracellular asparagine as a nitrogen source.
The hexose transporter family consists of 20 putative HXT genes (HXT1-HXT17, GAL2, SNF3, and RGT2) located on different chromosomes (Boles and Hollenberg 1997; Kruckeberg 1996) of which HXT15, HXT16 and HXT2 are absent from S. jurei. Under normal conditions, only 6 HXT genes (HXT1 and HXT3-HXT7) are known to play role in glucose uptake suggesting that loss of 3 HXT genes from S. jurei is unlikely to affect glucose transport (Lin and Li 2011).
Heterozygosis and strain divergence in the S. jurei
To detect genetic divergence between the two strains we mapped SNPs between the strains (NCYC 3947T vs. NCYC 3962), while to detect heterozygosis, we mapped the Single Nucleotide Polymorphisms (SNPs) in the two sets of alleles within the novel strains (NCYC 3947T vs. NCYC 3947T, and NCYC 3962 vs. NCYC 3962). We found 6227 SNPs between the two strains, showing a genetic divergence between them, which is relatively lower compared to the genetic divergence found among S. cerevisiae strains. Moreover, 278 and 245 SNPs were found within NCYC 3947T and NCYC 3962 strains respectively, indicating a low level of heterozygosity within each strain (Table 8). 139 SNPs were found be to common to both strains. Previous studies on S. cerevisiae and S. paradoxus strains from different lineages have shown that the level of heterozygosity is variable, with a large number of strains showing high level of heterozygosity isolated from human associated environments (Magwene et al. 2011; Tsai et al. 2008). A more recent study on 1011 S. cerevisiae natural strains showed that 63% of the sequenced isolates were heterozygous (Peter et al. 2018).
Table 8. Approximate numbers of SNPs in S. jurei NCYC 3947T and NCYC 3962 genome.
| Reference genome | Genome mapped | SNPs |
|---|---|---|
| NCYC 3947T | NCYC 3947T | 278 |
| NCYC 3962 | NCYC 3962 | 245 |
| NCYC 3947T | NCYC 3962 | 5702 |
| NCYC 3962 | NCYC 3947T | 6227 |
Phylogenetic analysis
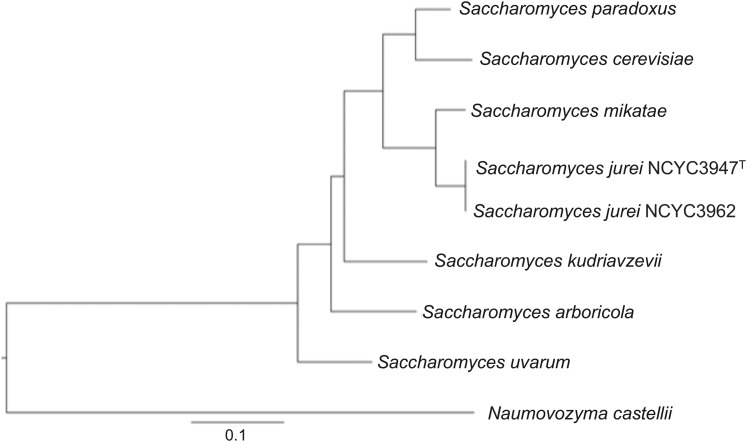
A first phylogeny construction using ITS/D1+D2 sequence analysis showed that S. jurei is placed in the tree close to S. cerevisiae, S. mikatae and S. paradoxus (Naseeb et al. 2017b). Here, we reconstructed the phylogeny using a multigene concatenation approach, which combines many genes together giving a large alignment (Fitzpatrick et al. 2006; Brown et al. 2001; Baldauf et al. 2000). Combination of concatenated genes improves the phylogenetic accuracy and helps to resolve the nodes and basal branching (Rokas et al. 2003). To reconstruct the evolutionary events, we concatenated 101 universally distributed orthologs obtained from complete genome sequencing data (Table S7). Both novel strains were located in one single monophyletic group, with the S. mikatae (Figure 4). Since S. jurei also have a chromosomal translocation in common with S. mikatae, it further shows that the two species share similar evolutionary history and hence present in the same group on the phylogenetic tree.
Figure 4.
Phylogenetic tree showing both novel strains located in one single monophyletic group, with the S. mikatae. Maximum likelihood phylogeny was constructed using a concatenated alignment of 101 universally distributed genes. Sequences from all Saccharomyces sensu stricto species were aligned using StatAlign v3.1 and phylogenetic tree was built using RaxML 8.1.3 with N. castellii kept as out-group.
Introgression analysis
To determine whether the two S. jurei strains possessed any introgressed region from other yeast species, we compared S. jurei genome with those of S. cerevisiae, S. mikatae, S. paradoxus and S. kudriavzevii. We did not observe introgression of any full-length genes or large segments of the genome (>1000 bp) in S. jurei. However, in both novel strains, we identified seven small DNA fragments (300 bp-700 bp) belonging to five different genes, which may have derived from S. paradoxus or S. mikatae (Table S8). DNA fragments from all the genes (CSS3, IMA5, MAL33, YAL003W) with the exception of YDR541C, showed a high sequence similarity to S. paradoxus genome, indicating putative introgression from S. paradoxus to S. jurei (Table S8).
Introgression of genetic material can easily occur in Saccharomyces species by crossing the isolates to make intraspecific or interspecific hybrids (Fischer et al. 2000; Naumov et al. 2000). Among the Saccharomyces sensu stricto group, introgressions have been demonstrated in natural and clinical yeast isolates (Liti et al. 2006; Zhang et al. 2010; Wei et al. 2007; Muller and McCusker 2009) and in wine, beer and other fermentation environments (de Barros Lopes et al. 2002; Usher and Bond 2009; Dunn et al. 2012). It is generally believed that introgressed regions are retained, as they may be evolutionarily advantageous (Strope et al. 2015; Novo et al. 2009). Previous studies have demonstrated that introgression in S. cerevisiae is relatively common and a majority of the genes are derived from introgression with S. paradoxus (Strope et al. 2015; Warringer et al. 2011; Novo et al. 2009; Liti et al. 2006; Peter et al. 2018).
Phenotypic profiling of S. jurei
We performed large-scale phenotypic profiling under various stress conditions and at different temperatures to capture the fitness landscape of S. jurei (strains NCYC 3947T and NCYC 3962) relative to other Saccharomyces sensu stricto species. Type strains of all Saccharomyces sensu stricto species were used for fitness analysis. Colony size was taken as a proxy for fitness score (see methods). Generally the fitness of S. jurei NCYC 3962 in different environmental stressor conditions was higher compared to S. jurei NCYC 3947T (Figure 3). Remarkably, only S. jurei NCYC 3962 was able to grow well on higher concentrations of acetic acid (Figure 3). Like most of the other Saccharomyces yeast species, both strains of S. jurei can also grow in media containing 10% ethanol. Although S. eubayanus showed the highest fitness in media containing 15% maltose, both strains of S. jurei were also able to tolerate high concentrations of maltose. Moreover, S. jurei NCYC 3962 was able to better tolerate higher concentrations of H2O2, CuSO4 and NaCl compared to most of the other sensu stricto species (Figure 3). Saccharomyces yeast species can acquire copper tolerance either due to an increase in CUP1 copy number (Warringer et al. 2011) or due to the use of copper sulfate as a fungicide in vineyards (Fay et al. 2004; Perez-Ortin et al. 2002). The genomic analysis shows that both strains of S. jurei possess one copy of CUP1, indicating other factors maybe associated with copper tolerance.
Phenotypically, both strains of S. jurei clustered with S. mikatae and S. paradoxus, which is in accordance with our phylogenetic results, and, interestingly, the brewing yeast S. eubayanus was also present in the same cluster (Figure 3). This may indicate that in spite of the phylogenetic distance, S. eubayanus may have shared similar ecological conditions with the other above mentioned species.
We also evaluated the fitness of S. jurei strains in comparison to Saccharomyces sensu stricto species at different temperatures, taking into account growth parameters such as lag phase (λ), maximum growth rate (µmax), and maximum biomass (Amax) (Tables S9-S11). The optimum growth of NCYC 3947T and NCYC 3962 was at 25° and 30° respectively (Table S10). Both strains of S. jurei are able to grow at a high temperatures (i.e., 37°) compared to S. kudriavzevii, S. pastorianus, S. arboricola, S. uvarum, and S. eubayanus, which are unable to grow at 37° (Table S10). The ability of S. jurei strains to grow well both at cold and warm suggest that this species evolved to be a generalist rather than a specialist in terms of thermoprofiles. The growth profiles captured at different temperatures for the other Saccharomyces species was in accordance to the previously published study (Salvadó et al. 2011).
Conclusions
High quality de novo assembly of two novel strains of S. jurei (NCYC 3947T and NCYC 3962) has been carried out using short and long reads sequencing strategies. We obtained a 12 Mb genome and were able to assemble full chromosomes of both strains. We found two reciprocal chromosomal translocations in S. jurei genome, between chromosomes I/XIII and VI/VII. The translocation between chromosomes I/XIII is unique to S. jurei genome, whereas the translocation between VI/VII is shared with S. mikatae IFO1815 and IFO1816. This suggests a common origin between S. jurei and S. mikatae and S. jurei evolved after acquiring the translocation between chromosomes I/XIII, while S. mikatae 1815 acquired a second translocation between chromosomes XVI/VII. Moreover, both strains of S. jurei showed low heterozygosis within themselves and were genetically diverged possessing 6227 SNPs between them. We found 4 novel ORFs that had no significant match to any of the available genomes. S. jurei genome had an increased number of Ty elements compared to S. cerevisiae and showed no signatures of introgression. The phylogenetic analysis showed that the novel species is closely related to S. mikatae, forming a single monophyletic group.
Phenotypically, the environmental stressor profiles of S. jurei are similar to those of with S. mikatae, S. paradoxus, S. cerevisiae (which further reiterate that S. jurei is closely related to these species) and S. eubayanus. We found that S. jurei NCYC 3962 compared to other sensu stricto species was able to grow well at high concentrations of acetic acid. In general, S. jurei NCYC 3962 showed relatively higher fitness compared to S. jurei NCYC 3947T under most of the environmental stress conditions tested. Both strains of S. jurei showed similar growth rate at relatively low temperature, however, NCYC 3962 showed increased fitness compared to NCYC 3947T at higher temperatures. The sequencing data and the large-scale phenotypic screening of this new species provide the basis for future investigations of biotechnological and industrial importance.
Acknowledgments
The authors would like to thank Genomic Technologies Core Facility at the University of Manchester for Illumina Hi-seq and Dr. Haiping Hao at Deep Sequencing and Microarray Core Facility of Johns Hopkins University for PacBio sequencing. SN is supported through BBSRC funding (BB/L021471/1). HA is supported by a scholarship funded by the Kuwait government through Kuwait University.
Footnotes
Supplemental material available at Figshare: https://figshare.com/s/60bbbc1e98886077182a.
Communicating editor: C. Boone
Literature Cited
- Adamczyk J., Deregowska A., Skoneczny M., Skoneczna A., Natkanska U., et al. , 2016. Copy number variations of genes involved in stress responses reflect the redox state and DNA damage in brewing yeasts. Cell Stress Chaperones 21: 849–864. 10.1007/s12192-016-0710-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo G. M., Lotti M., Tamas M. J., Brocca S., 2012. Amplification of the CUP1 gene is associated with evolution of copper tolerance in Saccharomyces cerevisiae. Microbiology 158: 2325–2335. 10.1099/mic.0.058024-0 [DOI] [PubMed] [Google Scholar]
- Adams J., Puskas-Rozsa S., Simlar J., Wilke C. M., 1992. Adaptation and major chromosomal changes in populations of Saccharomyces cerevisiae. Curr. Genet. 22: 13–19. 10.1007/BF00351736 [DOI] [PubMed] [Google Scholar]
- Avelar A. T., Perfeito L., Gordo I., Ferreira M. G., 2013. Genome architecture is a selectable trait that can be maintained by antagonistic pleiotropy. Nat. Commun. 4: 2235 10.1038/ncomms3235 [DOI] [PubMed] [Google Scholar]
- Baker E., Wang B., Bellora N., Peris D., Hulfachor A. B., et al. , 2015. The Genome Sequence of Saccharomyces eubayanus and the Domestication of Lager-Brewing Yeasts. Mol. Biol. Evol. 32: 2818–2831. 10.1093/molbev/msv168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf S. L., Roger A. J., Wenk-Siefert I., Doolittle W. F., 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290: 972–977. 10.1126/science.290.5493.972 [DOI] [PubMed] [Google Scholar]
- Bao W., Kojima K. K., Kohany O., 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6: 11 10.1186/s13100-015-0041-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund A. C., Sjolund E., Ostlund G., Sonnhammer E. L., 2008. InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Res. 36: D263–D266. 10.1093/nar/gkm1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström A., Simpson J. T., Salinas F., Barré B., Parts L., et al. , 2014. A high-definition view of functional genetic variation from natural yeast genomes. Mol. Biol. Evol. 31: 872–888. 10.1093/molbev/msu037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleykasten-Grosshans C., Friedrich A., Schacherer J., 2013. Genome-wide analysis of intraspecific transposon diversity in yeast. BMC Genomics 14: 399 10.1186/1471-2164-14-399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles E., Hollenberg C. P., 1997. The molecular genetics of hexose transport in yeasts. FEMS Microbiol. Rev. 21: 85–111. 10.1111/j.1574-6976.1997.tb00346.x [DOI] [PubMed] [Google Scholar]
- Bourque G., Zdobnov E. M., Bork P., Pevzner P. A., Tesler G., 2005. Comparative architectures of mammalian and chicken genomes reveal highly variable rates of genomic rearrangements across different lineages. Genome Res. 15: 98–110. 10.1101/gr.3002305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice C., Cubillos F. A., Dequin S., Camarasa C., Martinez C., 2018. Adaptability of the Saccharomyces cerevisiae yeasts to wine fermentation conditions relies on their strong ability to consume nitrogen. PLoS One 13: e0192383 10.1371/journal.pone.0192383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridier-Nahmias A., Tchalikian-Cosson A., Baller J. A., Menouni R., Fayol H., et al. , 2015. Retrotransposons. An RNA polymerase III subunit determines sites of retrotransposon integration. Science 348: 585–588. 10.1126/science.1259114 [DOI] [PubMed] [Google Scholar]
- Brown J. R., Douady C. J., Italia M. J., Marshall W. E., Stanhope M. J., 2001. Universal trees based on large combined protein sequence data sets. Nat. Genet. 28: 281–285. 10.1038/90129 [DOI] [PubMed] [Google Scholar]
- Byrne K. P., Wolfe K. H., 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15: 1456–1461. 10.1101/gr.3672305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinali G., Martini A., 1994. Electrophoretic karyotypes of authentic strains of the sensu stricto group of the genus Saccharomyces. Int. J. Syst. Bacteriol. 44: 791–797. 10.1099/00207713-44-4-791 [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V., 1985. An electrophoretic karyotype for yeast. Proc. Natl. Acad. Sci. USA 82: 3756–3760. 10.1073/pnas.82.11.3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaregola S., Neuveglise C., Lepingle A., Bon E., Feynerol C., et al. , 2000. Genomic exploration of the hemiascomycetous yeasts: 17. Yarrowia lipolytica. FEBS Lett. 487: 95–100. 10.1016/S0014-5793(00)02288-2 [DOI] [PubMed] [Google Scholar]
- Challis D., Yu J., Evani U. S., Jackson A. R., Paithankar S., et al. , 2012. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics 13: 8 10.1186/1471-2105-13-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. E., Kolodner R. D., 2011. A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements. PLoS Genet. 7: e1002089 10.1371/journal.pgen.1002089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S. L., Lai H. Y., Tung S. Y., Leu J. Y., 2013. Dynamic large-scale chromosomal rearrangements fuel rapid adaptation in yeast populations. PLoS Genet. 9: e1003232 10.1371/journal.pgen.1003232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. S., Alexander D. H., Marks P., Klammer A. A., Drake J., et al. , 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10: 563–569. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., et al. , 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301: 71–76. 10.1126/science.1084337 [DOI] [PubMed] [Google Scholar]
- Colson I., Delneri D., Oliver S. G., 2004. Effects of reciprocal chromosomal translocations on the fitness of Saccharomyces cerevisiae. EMBO Rep. 5: 392–398. 10.1038/sj.embor.7400123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Barros Lopes M., Bellon J. R., Shirley N. J., Ganter P. F., 2002. Evidence for multiple interspecific hybridization in Saccharomyces sensu stricto species. FEMS Yeast Res. 1: 323–331. 10.1111/j.1567-1364.2002.tb00051.x [DOI] [PubMed] [Google Scholar]
- Delneri D., Gardner D. C., Bruschi C. V., Oliver S. G., 1999a Disruption of seven hypothetical aryl alcohol dehydrogenase genes from Saccharomyces cerevisiae and construction of a multiple knock-out strain. Yeast 15: 1681–1689. [DOI] [PubMed] [Google Scholar]
- Delneri D., Gardner D. C., Oliver S. G., 1999b Analysis of the seven-member AAD gene set demonstrates that genetic redundancy in yeast may be more apparent than real. Genetics 153: 1591–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham M. J., Badrane H., Ferea T., Adams J., Brown P. O., et al. , 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 16144–16149. 10.1073/pnas.242624799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn B., Richter C., Kvitek D. J., Pugh T., Sherlock G., 2012. Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res. 22: 908–924. 10.1101/gr.130310.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel S. R., Cherry J. M., 2013. The new modern era of yeast genomics: community sequencing and the resulting annotation of multiple Saccharomyces cerevisiae strains at the Saccharomyces Genome Database. Database (Oxford) 2013: bat012 10.1093/database/bat012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., McCullough H. L., Sniegowski P. D., Eisen M. B., 2004. Population genetic variation in gene expression is associated with phenotypic variation in Saccharomyces cerevisiae. Genome Biol. 5: R26 10.1186/gb-2004-5-4-r26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G., James S. A., Roberts I. N., Oliver S. G., Louis E. J., 2000. Chromosomal evolution in Saccharomyces. Nature 405: 451–454. 10.1038/35013058 [DOI] [PubMed] [Google Scholar]
- Fischer G., Neuveglise C., Durrens P., Gaillardin C., Dujon B., 2001. Evolution of gene order in the genomes of two related yeast species. Genome Res. 11: 2009–2019. 10.1101/gr.212701 [DOI] [PubMed] [Google Scholar]
- Fischer G., Rocha E. P., Brunet F., Vergassola M., Dujon B., 2006. Highly variable rates of genome rearrangements between hemiascomycetous yeast lineages. PLoS Genet. 2: e32 10.1371/journal.pgen.0020032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick D. A., Logue M. E., Stajich J. E., Butler G., 2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 6: 99 10.1186/1471-2148-6-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J. A., Huang J. C., Pukkila-Worley R., Alspaugh J. A., Mitchell T. G., et al. , 2005. Chromosomal translocation and segmental duplication in Cryptococcus neoformans. Eukaryot. Cell 4: 401–406. 10.1128/EC.4.2.401-406.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S., Hashimoto T., 2000. DNA fingerprinting patterns of Candida species using HinfI endonuclease. Int. J. Syst. Evol. Microbiol. 50: 1381–1389. 10.1099/00207713-50-3-1381 [DOI] [PubMed] [Google Scholar]
- Gertz E. M., Yu Y. K., Agarwala R., Schaffer A. A., Altschul S. F., 2006. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 4: 41 10.1186/1741-7007-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard M. R., Greig D., 2015. Saccharomyces cerevisiae: a nomadic yeast with no niche? FEMS Yeast Res. 15 10.1093/femsyr/fov009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., et al. , 1996. Life with 6000 genes. Science 274 (5287):546, 563–547. 10.1126/science.274.5287.546 [DOI] [PubMed] [Google Scholar]
- Gordon J. L., Byrne K. P., Wolfe K. H., 2009. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 5: e1000485 10.1371/journal.pgen.1000485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorter de Vries A. R., Pronk J. T., Daran J. G., 2017. Industrial Relevance of Chromosomal Copy Number Variation in Saccharomyces Yeasts. Appl. Environ. Microbiol. 83: e03206-16. 10.1128/AEM.03206-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C., Brachat S., Dietrich F. S., 2005. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot. Cell 4: 1102–1115. 10.1128/EC.4.6.1102-1115.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt S. K., Donaldson I. J., Lovell S. C., Delneri D., 2014. Sequencing and characterisation of rearrangements in three S. pastorianus strains reveals the presence of chimeric genes and gives evidence of breakpoint reuse. PLoS One 9: e92203 10.1371/journal.pone.0092203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Friedrich A., de Montigny J., Schacherer J., 2014. Chromosomal rearrangements as a major mechanism in the onset of reproductive isolation in Saccharomyces cerevisiae. Curr. Biol. 24: 1153–1159. 10.1016/j.cub.2014.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H. Y., Fraser M., Li W., et al. , 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhten P., Ponomarova O., Gonzalez R., Patil K. R., 2016. Saccharomyces cerevisiae metabolism in ecological context. FEMS Yeast Res. 16 10.1093/femsyr/fow080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S., 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254. 10.1038/nature01644 [DOI] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., et al. , 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruckeberg A. L., 1996. The hexose transporter family of Saccharomyces cerevisiae. Arch. Microbiol. 166: 283–292. 10.1007/s002030050385 [DOI] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A. L., Smoot M., Shumway M., et al. , 2004. Versatile and open software for comparing large genomes. Genome Biol. 5: R12 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry C. R., Oh J., Hartl D. L., Cavalieri D., 2006. Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene 366: 343–351. 10.1016/j.gene.2005.10.042 [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin D. M., Pape G., Donthu R., Auvil L., Welge M., et al. , 2009. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res. 19: 770–777. 10.1101/gr.086546.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- League G. P., Slot J. C., Rokas A., 2012. The ASP3 locus in Saccharomyces cerevisiae originated by horizontal gene transfer from Wickerhamomyces. FEMS Yeast Res. 12: 859–863. 10.1111/j.1567-1364.2012.00828.x [DOI] [PubMed] [Google Scholar]
- Lewis, S.E., S.M. Searle, N. Harris, M. Gibson, V. Lyer et al., 2002 Apollo: a sequence annotation editor. Genome Biol 3 (12):RESEARCH0082. [DOI] [PMC free article] [PubMed]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libkind D., Hittinger C. T., Valerio E., Goncalves C., Dover J., et al. , 2011. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. USA 108: 14539–14544. 10.1073/pnas.1105430108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Li W. H., 2011. Expansion of hexose transporter genes was associated with the evolution of aerobic fermentation in yeasts. Mol. Biol. Evol. 28: 131–142. 10.1093/molbev/msq184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Barton D. B., Louis E. J., 2006. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics 174: 839–850. 10.1534/genetics.106.062166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., et al. , 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341. 10.1038/nature07743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Nguyen Ba A. N., Blythe M., Muller C. A., Bergstrom A., et al. , 2013. High quality de novo sequencing and assembly of the Saccharomyces arboricolus genome. BMC Genomics 14: 69 10.1186/1471-2164-14-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Eddy S. R., 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. 10.1093/nar/25.5.0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2002. Genomics. Gene duplication and evolution. Science 297: 945–947. 10.1126/science.1075472 [DOI] [PubMed] [Google Scholar]
- Magwene P. M., Kayikci O., Granek J. A., Reininga J. M., Scholl Z., et al. , 2011. Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 108: 1987–1992. 10.1073/pnas.1012544108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini A. V., Kurtzman C. P., 1985. Deoxyribonucleic Acid Relatedness among Species of the Genus Saccharomyces Sensu Stricto. Int. J. Syst. Evol. Microbiol. 35: 508–511. [Google Scholar]
- Martini A. V., Martini A., 1987. Three newly delimited species of Saccharomyces sensu stricto. Antonie van Leeuwenhoek 53: 77–84. 10.1007/BF00419503 [DOI] [PubMed] [Google Scholar]
- Masneuf I., Hansen J., Groth C., Piskur J., Dubourdieu D., 1998. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl. Environ. Microbiol. 64: 3887–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski P. A., Lemoine F. J., Petes T. D., 2006. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst.) 5: 1010–1020. 10.1016/j.dnarep.2006.05.027 [DOI] [PubMed] [Google Scholar]
- Muller L. A., McCusker J. H., 2009. A multispecies-based taxonomic microarray reveals interspecies hybridization and introgression in Saccharomyces cerevisiae. FEMS Yeast Res. 9: 143–152. 10.1111/j.1567-1364.2008.00464.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W. J., Larkin D. M., Everts-van der Wind A., Bourque G., Tesler G., et al. , 2005. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309: 613–617. 10.1126/science.1111387 [DOI] [PubMed] [Google Scholar]
- Naseeb S., Ames R. M., Delneri D., Lovell S. C., 2017a Rapid functional and evolutionary changes follow gene duplication in yeast. Proc. Biol. Sci. 284 10.1098/rspb.2017.1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseeb S., Carter Z., Minnis D., Donaldson I., Zeef L., et al. , 2016. Widespread impact of chromosomal inversions on gene expression uncovers robustness via phenotypic buffering. Mol. Biol. Evol. 33: 1679–1696. 10.1093/molbev/msw045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseeb S., Delneri D., 2012. Impact of chromosomal inversions on the yeast DAL cluster. PLoS One 7: e42022 10.1371/journal.pone.0042022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseeb S., James S. A., Alsammar H., Michaels C. J., Gini B., et al. , 2017b Saccharomyces jurei sp. nov., isolation and genetic identification of a novel yeast species from Quercus robur. Int. J. Syst. Evol. Microbiol. 67: 2046–2052. 10.1099/ijsem.0.002013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov G. I., 1987. Genetic basis for classification and identification of the ascomycetous yeasts. Stud. Mycol. 30: 469–475. [Google Scholar]
- Naumov G. I., James S. A., Naumova E. S., Louis E. J., Roberts I. N., 2000. Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int. J. Syst. Evol. Microbiol. 50: 1931–1942. 10.1099/00207713-50-5-1931 [DOI] [PubMed] [Google Scholar]
- Naumov G. I., Naumova E. S., Hagler A. N., Mendonca-Hagler L. C., Louis E. J., 1995a A new genetically isolated population of the Saccharomyces sensu stricto complex from Brazil. Antonie van Leeuwenhoek 67: 351–355. 10.1007/BF00872934 [DOI] [PubMed] [Google Scholar]
- Naumov G. I., Naumova E. S., Louis E. J., 1995b Two new genetically isolated populations of the Saccharomyces sensu stricto complex from Japan. J. Gen. Appl. Microbiol. 41: 499–505. 10.2323/jgam.41.499 [DOI] [PubMed] [Google Scholar]
- Naumov G. I., Naumova E. S., Sancho E. D., 1996. Genetic reidentification of Saccharomyces strains associated with black knot disease of trees in Ontario and Drosophila species in California. Can. J. Microbiol. 42: 335–339. 10.1139/m96-049 [DOI] [Google Scholar]
- Nawrocki E. P., Burge S. W., Bateman A., Daub J., Eberhardt R. Y., et al. , 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43: D130–D137. 10.1093/nar/gku1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki E. P., Eddy S. R., 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29: 2933–2935. 10.1093/bioinformatics/btt509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. V., Legras J. L., Neuveglise C., Gaillardin C., 2011. Deciphering the hybridisation history leading to the Lager lineage based on the mosaic genomes of Saccharomyces bayanus strains NBRC1948 and CBS380. PLoS One 6: e25821 10.1371/journal.pone.0025821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo M., Bigey F., Beyne E., Galeote V., Gavory F., et al. , 2009. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. USA 106: 16333–16338. 10.1073/pnas.0904673106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S., Johnston M., 1999. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 63: 554–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Ortin J. E., Querol A., Puig S., Barrio E., 2002. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 12: 1533–1539. 10.1101/gr.436602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter J., De Chiara M., Friedrich A., Yue J. X., Pflieger D., et al. , 2018. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556: 339–344. 10.1038/s41586-018-0030-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvirenti A., Nguyen H., Caggia C., Giudici P., Rainieri S., et al. , 2000. Saccharomyces uvarum, a proper species within Saccharomyces sensu stricto. FEMS. Microbiol. Lett. 192: 191–196. [DOI] [PubMed] [Google Scholar]
- Querol A., Bond U., 2009. The complex and dynamic genomes of industrial yeasts. FEMS Microbiol. Lett. 293: 1–10. 10.1111/j.1574-6968.2008.01480.x [DOI] [PubMed] [Google Scholar]
- Raney B. J., Dreszer T. R., Barber G. P., Clawson H., Fujita P. A., et al. , 2014. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics 30: 1003–1005. 10.1093/bioinformatics/btt637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A., Williams B. L., King N., Carroll S. B., 2003. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425: 798–804. 10.1038/nature02053 [DOI] [PubMed] [Google Scholar]
- Salvadó Z., Arroyo-López F. N., Guillamón J. M., Salazar G., Querol A., et al. , 2011. Temperature adaptation markedly determines evolution within the genus Saccharomyces. Appl. Environ. Microbiol. 77: 2292–2302. 10.1128/AEM.01861-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell D. R., Zill O. A., Rokas A., Payen C., Dunham M. J., et al. , 2011. The awesome power of yeast evolutionary genetics: New genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3 (Bethesda) 1: 11–25. 10.1534/g3.111.000273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C., Federspiel N., Jones T., Hansen N., Bivolarovic V., et al. , 2000. Prevalence of small inversions in yeast gene order evolution. Proc. Natl. Acad. Sci. USA 97: 14433–14437. 10.1073/pnas.240462997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y., Malhotra A., Bekiranov S., Dutta A., 2009. Yeast genome analysis identifies chromosomal translocation, gene conversion events and several sites of Ty element insertion. Nucleic Acids Res. 37: 6454–6465. 10.1093/nar/gkp650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A. F., Hubley A., R., Green P., 2013–2015. RepeatMasker Open-4.0. http://www.repeatmasker.org.
- Soares E. V., 2011. Flocculation in Saccharomyces cerevisiae: a review. J. Appl. Microbiol. 110: 1–18. 10.1111/j.1365-2672.2010.04897.x [DOI] [PubMed] [Google Scholar]
- Stanke, M., and B. Morgenstern, 2005 AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33 (Web Server issue):W465–467. 10.1093/nar/gki458. 10.1093/nar/gki458 [DOI] [PMC free article] [PubMed]
- Strope P. K., Skelly D. A., Kozmin S. G., Mahadevan G., Stone E. A., et al. , 2015. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 25: 762–774. 10.1101/gr.185538.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai I. J., Bensasson D., Burt A., Koufopanou V., 2008. Population genomics of the wild yeast Saccharomyces paradoxus: Quantifying the life cycle. Proc. Natl. Acad. Sci. USA 105: 4957–4962. 10.1073/pnas.0707314105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usher J., Bond U., 2009. Recombination between homoeologous chromosomes of lager yeasts leads to loss of function of the hybrid GPH1 gene. Appl. Environ. Microbiol. 75: 4573–4579. 10.1128/AEM.00351-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakirlis N., Sarilar V., Drillon G., Fleiss A., Agier N., et al. , 2016. Reconstruction of ancestral chromosome architecture and gene repertoire reveals principles of genome evolution in a model yeast genus. Genome Res. 26: 918–932. 10.1101/gr.204420.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther A., Hesselbart A., Wendland J., 2014. Genome sequence of Saccharomyces carlsbergensis, the world’s first pure culture lager yeast. G3 (Bethesda) 4: 783–793. 10.1534/g3.113.010090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. A., Bai F. Y., 2008. Saccharomyces arboricolus sp. nov., a yeast species from tree bark. Int. J. Syst. Evol. Microbiol. 58: 510–514. 10.1099/ijs.0.65331-0 [DOI] [PubMed] [Google Scholar]
- Warringer J., Zorgo E., Cubillos F. A., Zia A., Gjuvsland A., et al. , 2011. Trait variation in yeast is defined by population history. PLoS Genet. 7: e1002111 10.1371/journal.pgen.1002111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., McCusker J. H., Hyman R. W., Jones T., Ning Y., et al. , 2007. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc. Natl. Acad. Sci. USA 104: 12825–12830. 10.1073/pnas.0701291104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Mikata K., Banno I., 1993. Reidentification of 121 strains of the genus Saccharomyces. Bull. Jpn. Fed. Cult. Coll. 9: 95–119. [Google Scholar]
- Zhang H., Skelton A., Gardner R. C., Goddard M. R., 2010. Saccharomyces paradoxus and Saccharomyces cerevisiae reside on oak trees in New Zealand: evidence for migration from Europe and interspecies hybrids. FEMS Yeast Res. 10: 941–947. 10.1111/j.1567-1364.2010.00681.x [DOI] [PubMed] [Google Scholar]