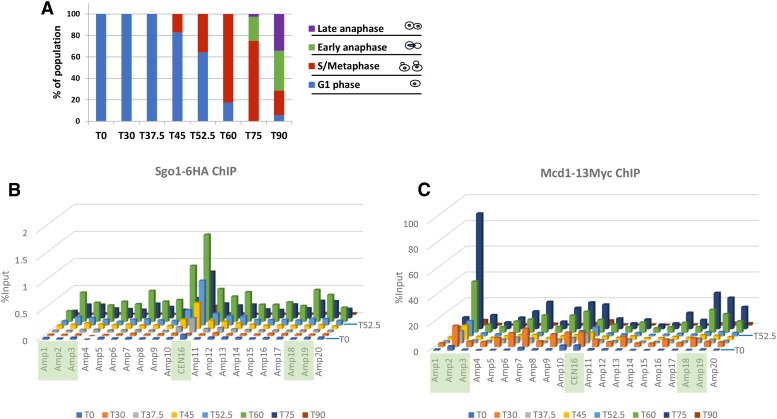
Figure 5.
Dynamic recruitment of Sgo1p and cohesin at centromere and pericentromere through cell cycle. A. Budding index of cells collected from the indicated time points. B and C. Sgo1p-HA and Mcd1p-Myc co-expressed in the same cells were examined by ChIP-qPCR. PCR amplicons correspond to CEN16 and nearby regions. See Figure 3 for positions of these amplicons. In addition to the conspicuous difference in the IP efficiencies between Mcd1p-Myc and Sgo1p-HA (also see Figure 3), we noticed that the ChIP efficiency (%IP) of both Sgo1-6HA and Mcd1-13Myc in cells synchronously progressing through cell cycle was consistently 5- to 10-fold higher than in those benomyl-arrested cells. Results shown in this figure are a representative of two biological replicas. In both cases, the Mcd1p-13Myc exhibited significantly higher IP efficiency.