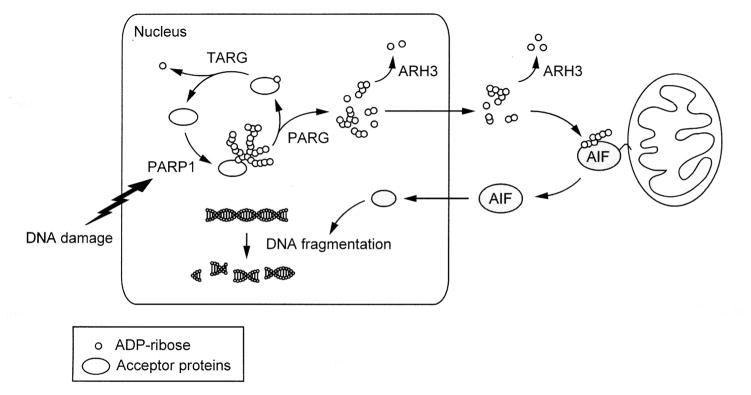
Fig. 1. ARH3 participates in PAR degradation in the nucleus and cytoplasm, thereby inhibiting parthanatos.
Upon DNA damage, activated PARP1 synthesizes PAR from NAD+, resulting in the poly-ADP-ribosylation of nuclear acceptor proteins including PARP1 itself. 110-kDa nuclear PARG, through its exo- and endo-glycosidase activities, degrades PAR polymers, generating ADP-ribose monomers and small PAR fragments, respectively. The initial ADP-ribose attached to acceptor proteins is degraded by macrodomain-containing proteins such as TARG1 (terminal ADP-ribose glycohydrolase; C6orf130) which are responsible for release of an ADP-ribose monomer attached to protein [78]. Small PAR fragments then pass through nuclear membranes, although the detailed mechanisms are still unknown. In the cytoplasm, PAR binds to the PAR-binding site AIF, releasing AIF from mitochondrial membranes. Released AIF is translocated to the nucleus, resulting in DNA fragmentation and cell death. ARH3 is located in the nucleus and cytoplasm, where ARH3 degrades PAR, preventing AIF release from mitochondria.