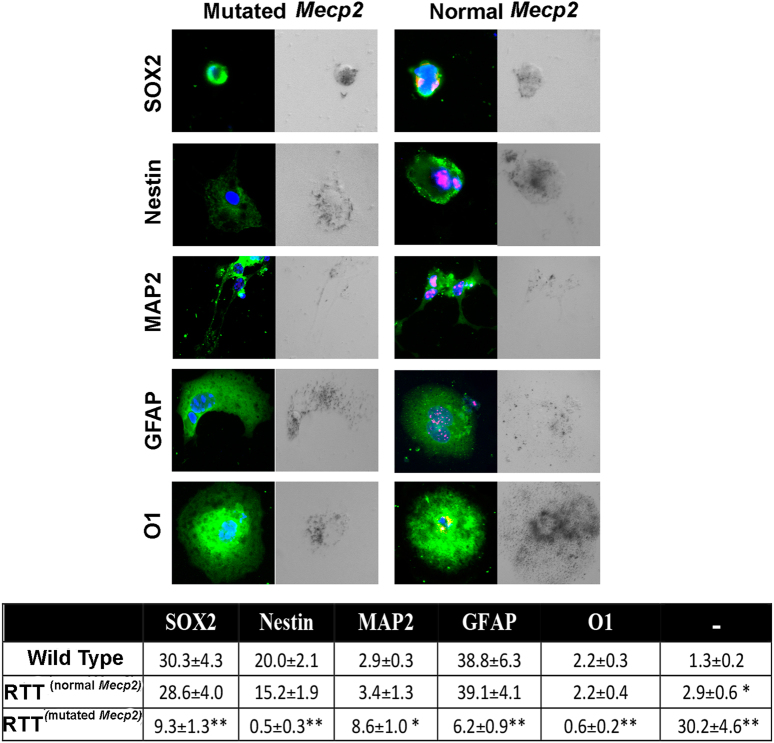
Fig. 5. NSC differentiation and senescence in cells expressing wild-type and mutated Mecp2.
Cells expressing mutated or wild-type Mecp2 were identified by using the anti-MECP2 primary antibody. In Mecp2+/− samples, the MECP2 protein (red) was present in cells expressing the wild-type allele and was undetectable in those expressing the mutated allele. The picture shows representative photomicrographs of Mecp2+/− and control NSCs differentiated for 3 days onto poly-l-ornithine-coated plates in medium without growth factors. The cells were stained in green for SOX2, Nestin, MAP2, GFAP, and O1. The nuclei were counterstained with DAPI (blue). Every microscopic field was also analyzed under a bright-light field to detect senescent-associated β-galactosidase. The senescent cells show a dark gray staining. In the table are indicated the percentages of stem cells (SOX2+), early progenitors (Nestin+), neurons (MAP2+), astrocytes (GFAP+), and oligodendrocytes (O1+); the (−) symbol indicates the cells that were negative for all the analyzed markers. In the Mecp2+/− samples, indicated as RTT, the percentage of progenitors and differentiated cells was evaluated in cells expressing either the normal or the mutated allele (n = 5 biological replicates; *p < 0.05, **p < 0.01 with respect to samples from wild-type animals)