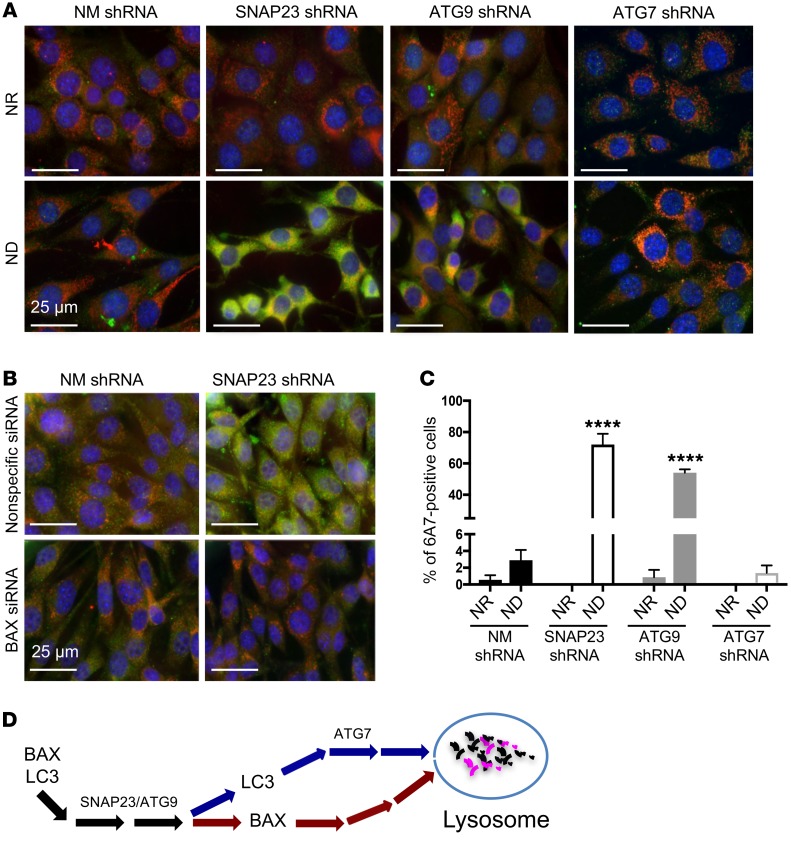
Figure 7. SNAP23 and ATG9 deficiency enhances BAX activation by ND.
(A) NM shRNA, SNAP23 shRNA, ATG9 shRNA, and ATG7 shRNA NIH3T3 cells were maintained under NR or ND conditions for 1 hour. Cells were fixed and subjected to immunofluorescence microscopy using the BAX activation–specific monoclonal antibody 6A7 (green), the mitochondria-specific antibody ATP5α (red), and DAPI (blue). Images are representative of 4 independent determinations. Scale bars: 25 μm. (B) Nonspecific siRNA and BAX siRNA–knockdown cells in the context of control NM shRNA and SNAP23 shRNA cells were maintained under ND conditions for 1 hour. Cells were fixed and subjected to immunofluorescence microscopy using the BAX activation–specific monoclonal antibody 6A7 (green), the mitochondria-specific antibody ATP5α (red), and DAPI (blue). Images are representative of 3 independent determinations. Scale bars: 25 μm. (C) Quantification of 6A7-positive cells from A was determined by counting 500 cells for each condition from 4 independent experiments. ****P < 0.0001, by ANOVA with Tukey’s post hoc test. Data represent the mean ± SEM. (D) Schematic model depicting a selective SNAP23/ATG9-dependent, but ATG7-independent, pathway mediating lysosomal degradation of BAX.