Fig. 1.
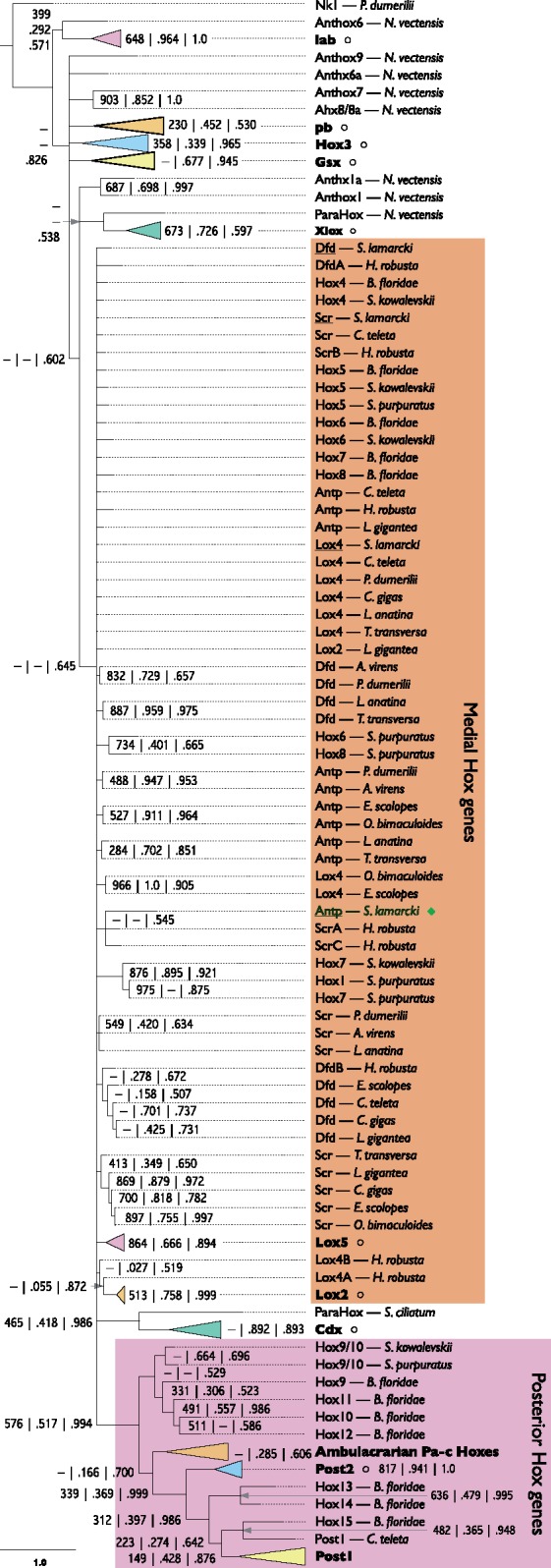
—Bayesian phylogeny of Hox and ParaHox homeodomains and flanking sequences from a selection of metazoan genomes, showing the basis for the identification of the divergent Spirobranchus Hox gene as Antp. Support values for each node are from neighbor-joining (out of 1000 bootstraps), maximum likelihood (proportion of 1000 bootstraps), and Bayesian (posterior probability) phylogenies (in order, separated by vertical bars or newlines). A dash indicates where a node is not present in the corresponding tree. Gene families that have been successfully reconstructed have been collapsed into colored triangles and a summary of their contents given in supplementary file 1, Supplementary Material online. Spirobranchus sequences (all underlined) are marked with a green diamond if found in the regenerative transcriptomes, and with a black circle if only found in the genome (collapsed families only). The scale bar indicates amino acid substitutions per site. Full sequence details are included in supplementary file 1, Sheet 2, Supplementary Material online. The original alignment is presented in supplementary file 7, Supplementary Material online. A full version of the Newick format tree is presented in supplementary file 2, Supplementary Material online. Annelid species: S. lamarcki, Spirobranchus lamarcki; C. teleta, Capitella teleta; A. virens, Alitta virens; H. robusta, Helobdella robusta; P. dumerilii, Platynereis dumerilii. Brachiopod species: L. anatina, Lingula anatina; T. transversa, Terebratalia transversa. Mollusc species: C. gigas, Crassostrea gigas; L. gigantea, Lottia gigantea; E. scolopes, Euprymna scolopes; O. bimaculoides, Octopus bimaculoides. Deuterostome species: B. floridae, Branchiostoma floridae; S. kowalevski, Saccoglossus kowalevskii; S. purpuratus, Strongylocentrotus purpuratus. Cnidarian species: N. vectensis, Nematostella vectensis. Poriferan species: S. ciliatum, Sycon ciliatum.
