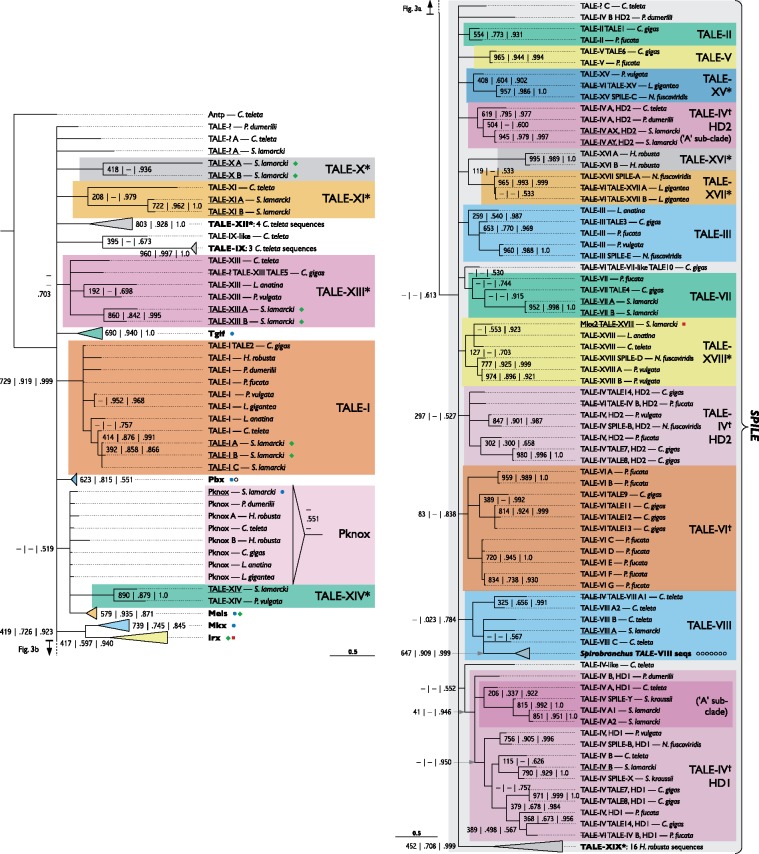
Fig. 3.
—Bayesian phylogeny of TALE class homeodomain sequences from a selection of lophotrochozoan genomes, showing the frequent duplication of canonical TALE class genes and the basis of our proposed revision to the TALE clade classification (Paps et al. 2015); split into two parts, a (left side) and b (right side). The SPILE clade (per Morino et al., 2018) is marked by a grey box and labelled bracket in part b. Support values and formatting as in figure 1. In some cases, new families or family subsets containing several sequences all from a single genus have also been collapsed to aid visualization. Single genus families are highlighted in grey, but otherwise color selection is arbitrary, and not meant to indicate a relationship except in the case of the TALE-IV clades. Similarly, paralogue lettering, where present, is not intended to consistently imply direct orthology, though where evident, direct orthologues have been lettered accordingly. S. lamarcki sequences (all underlined) are marked with a green diamond if found in the regenerative transcriptomes, with a red square if found in the developmental transcriptome (Kenny and Shimeld 2012), and a blue dot if found in both. Collapsed families have their S. lamarcki gene complement indicated nearby with the same symbols as above, with an open circle indicating a gene that has been found only in the genome. New gene families suggested herein are marked with an asterisk. Gene families that have gained or lost sequences from Paps et al. (2015) are marked with a dagger. Where a gene has been reclassified from Paps et al. (2015) or Kenny and Shimeld (2012), the old classification is included but struck out. Established gene families that were successfully reconstructed in the neighbor-joining and/or maximum likelihood analyses but not the Bayesian analysis are marked by a “cartoon” clade (not to horizontal scale) and corresponding support values to the right-hand side. The scale bar indicates amino acid substitutions per site. Full sequence details are included in supplementary file 1, Sheet 5, Supplementary Material online. The original alignment is presented in supplementary file 8, Supplementary Material online. A full version of the Newick format tree is presented in supplementary file 3, Supplementary Material online. Annelid species: S. lamarcki, Spirobranchus lamarcki; S. kraussi, Spirobranchus (formerly Pomatoleios) kraussi; C. teleta, Capitella teleta; H. robusta, Helobdella robusta; P. dumerilii, Platynereis dumerilii. Brachiopod species: L. anatina, Lingula anatina. Mollusc species: C. gigas, Crassostrea gigas; P. fucata, Pinctada fucata; L. gigantea, Lottia gigantea; N. fuscoviridis, Nipponacmea fuscoviridis; P. vulgata, Patella vulgata. Insect species (only in collapsed clades): Tribolium castaneum; Drosophila melanogaster.