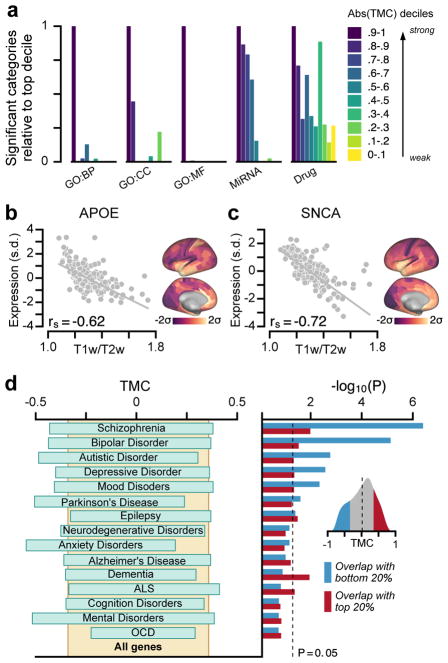
Figure 8.
Hierarchical variation relates to enrichment in neurobiological function and brain disorders. (a) Genes with strong TMCs are overrepresented in functional annotations across multiple gene ontologies (GOs). BP, biological process; CC, cellular component; MF, molecular function; MiRNA, microRNA binding sites; Drug, drug targets. (b, c) Two key risk genes for neurodegenerative disorders, APOE for Alzheimer’s disease and SNCA for Parkinson’s disease, exhibit strongly negative TMCs, with higher expression levels in association cortex relative to sensory cortex (APOE: rs = −0.62, P < 10−5; SNCA: rs = −0.72, P < 10−5; Spearman rank correlation). APOE is a leading risk gene for Alzheimer’s disease. The ε4 allele of APOE is the largest genetic risk factor for late-onset Alzheimer’s disease. SNCA (PARK1/PARK4) is a key risk gene for Parkinson’s disease. Duplication of SNCA is risk factor for familial Parkinson’s disease with dominant inheritance. SNCA codes for the alpha-synuclein protein which is the primary component of Lewy bodies, a biomarker of Parkinson’s disease. (d) Genes with strong negative TMCs are overrepresented in multiple gene sets associated with neuropsychiatric disorders. Left: 20–80% percentile range of TMCs for each disease gene set. Right: Enrichment is quantified by the hypergeometric test, which assesses the statistical significance of overlap between each gene set and the top (red) or bottom (blue) 20% TMC genes. Inset: Distribution of TMCs across all genes. Expression in panels b and c is plotted in units of standard deviations (s.d.; σ) from the mean for each map.