Figure 5. Haplotype aware genome editing of a Mendelian disease SNP in FLCN demonstrates that expression regulatory variation can modify its penetrance.
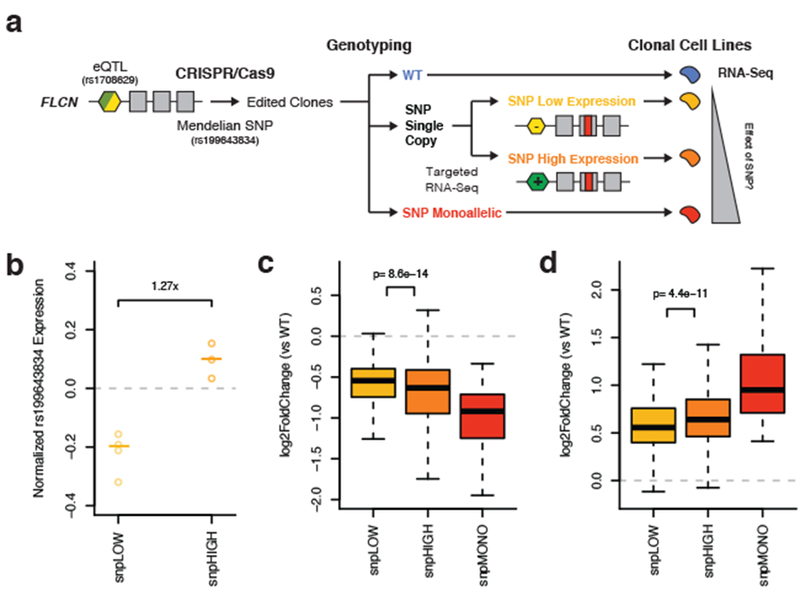
a) Illustration of the experimental study design. Briefly, a SNP that causes Birt-Hogg-Dubé Syndrome was edited on distinct eQTL haplotypes in 293T cells using CRISPR/Cas9. Monoclonal cell lines were genotyped and classified as monoallelic for the edit SNP (snpMONO), or as having a single copy. Using targeted RNA-seq, single copy clones were classified as either those where the edited SNP was on the lower (snpLOW) or higher (snpHIGH) expressed haplotype. The global transcriptome was used a cellular phenotype to assess SNP penetrance. b) Copy number normalized expression of the edited SNP as measured by targeted RNA-seq (allelic expression, log2(ALT/REF)) in snpLOW (allelic expression < 0, p-value < 0.01, binomial test versus 0.5) and snpHIGH (allelic expression > 0, p-value < 0.01) clones. Center lines represent median. c-d) Change in expression of genes that were significantly downregulated (c, 277 genes) or upregulated (d, 387 genes) in clones monoallelic for the edited SNP versus wild-type controls. Single copy edit SNP clones are stratified by haplotype configuration. P-values were calculated using a two-sided paired Wilcoxon signed rank test. See Supplementary Figure 7 and Supplementary Tables 4-6 for experimental details and additional analyses. For all plots, N = 4 snpLOW, 3 snpHIGH, 2 snpMONO, and 4 WT biologically independent samples. For boxplots, bottom whisker: Q1–1.5*IQR, top whisker: Q3+1.5*IQR, box: IQR, center: median, and outliers are not plotted for ease of viewing.
