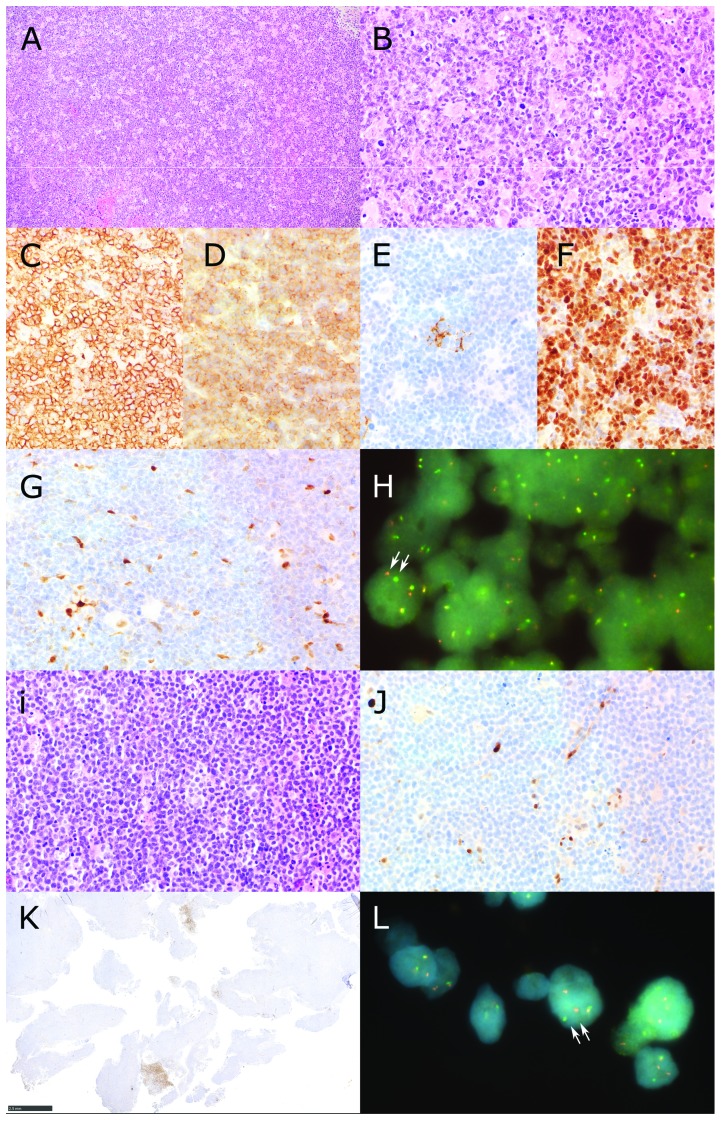
Figure 1.
Identification of CCND1-negative MCL Patients. Patient A (panels A-H) showed a diffuse infiltrate of slightly pleomorphic centrocytes (A and B Hematoxilin and Eosin) with expression of CD20 (C), CD5 (D), absence of CD23 but scattered follicular dendritic cells (E) and expression of SOX11 (F). The lymphoma was high proliferative with Ki67 positivity up to 60%. Staining for Cyclin D1 (CCND1) revealed internal positive controls (macrophages and endothelial cells) but no staining of lymphoma cells (G). Fluorescence in situ hybridization using a break apart probe shows that the CCND1 gene of patient A has a chromosomal rearrangement (H). A cell nucleus where the segregation of the two fluorescent probes (red and green) is clearly visible is indicated by two arrows. Patient B (panels I-L) presented with a diffuse infiltrate with a classical cytology of mantle cell lymphoma (I, Hematoxilin and Eosin). There was expression of CD20, CD5 and SOX11 (data not shown). Also this patient presented a high proliferative lymphoma with Ki67 postivity of 70%. Expression of CCND1 was not detectable despite internal positive controls (macrophages and endothelial cells, J) except for the presence of small areas in the whole lymphomatous tissue containing CCND1 positive lymphatic cells with pleomorphic cytology (K). This could indicate intratumoral clonal heterogeneity in this lymphoma case affecting, for unknown reason, CCND1 reactivity. (L) Fluorescence in situ hybridization using a break apart probe shows that the CCND1 gene of patient B has a chromosomal rearrangement. A cell nucleus where the segregation of the two fluorescent probes (red and green) is clearly visible is indicated by two arrows (Original magnification A: 100×, B-G, i,J: 400×, H, L 1000×, K. 20×).