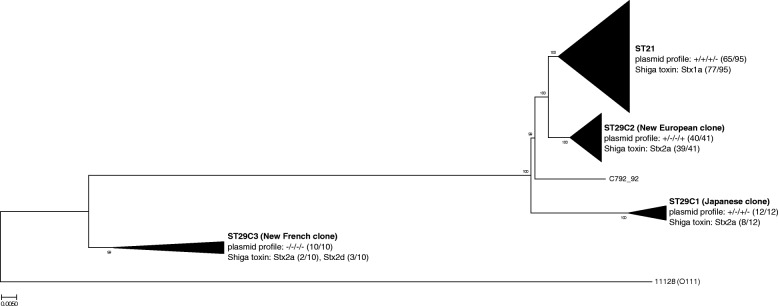
Fig. 1.
Global population structure of E. coli O26 based on whole genome phylogeny. The major lineages are named according to the current convention [9, 39]; strain C792_92 did not group within any lineage. The proportions of strains carrying plasmid gene profile characteristic for particular lineage (EHEC-hlyA/katP/espP/etpD) are denoted in parentheses. Predominant type of Shiga toxin and its proportion among strains of each particular lineage are denoted in parentheses. Core genome phylogeny was constructed from 12,205 variable bases among 3,718,610 validated homologous nucleotide positions. Phylogenetic tree was constructed with FastTree using approximately Maximum Likelihood phylogeny model [60]. Genomic sequence of E. coli O111:H− strain 11128 (stripped of plasmids, prophages and other horizontally acquired regions [68]) was used as an outgroup. For additional information about particular strains, see Additional file 1: Table S1. For a complete (unreduced) phylogram, see Additional file 2: Figure S1