Figure 2. High-resolution single-particle cryo-EM on GO grids.
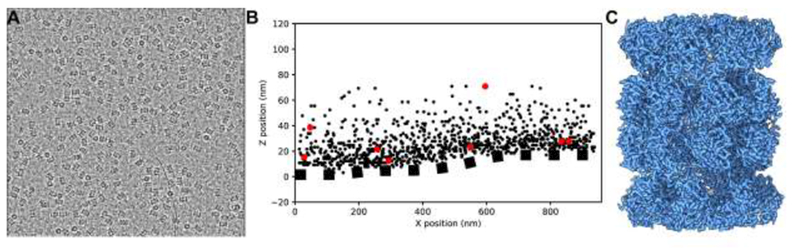
A. Micrograph of 20S proteasome particles over a GO support film. The GO film adds minimal background contrast. The specimen was applied at a concentration 0.05mg/mL, approximately ten times less than we typically use for this specimen. B. 3D localization of 20S particles from cryogenic electron tomography (cryo-ET). This tomogram was taken from a different GO grid than the one we used for the single-particle results: the 20S proteasome concentration here was higher and the GO substrate was far thicker (5-8 sheets). These conditions enabled us to identify how densely the GO surface could be coated with macromolecular specimen and to discern the position of the GO layer within low-resolution tomogram. Fiducial markers and points across the GO substrate surface were picked manually, while 20S proteasome particles were located in the tomogram using template matching. The GO is not flat but ripples and bends, as shown by the dotted line fitted by a spline to the points manually picked on the GO surface. The maximal distance between a picked proteasome particle and the GO substrate surface was 66nm, bounding the relative position of the air-water interface. 862 out of 969 picked proteasome particles (88.9%) are closer to the GO substrate than the air-water interface, strongly suggesting a direct interaction. All tomographic processing was performed using the IMOD software suite (Mastronarde, 2005). C. 3D reconstruction of the 20S proteasome on a GO support resolved at 2.5Å.
