Abstract
High-resolution capillary zone electrophoresis – mass spectrometry (CZE-MS) has been of increasing interest for the analysis of biopharmaceuticals. In this work, a combination of middle-down and intact CZE-MS analyses has been implemented for the characterization of a biotherapeutic monoclonal antibody (mAb) with a variety of post-translational modifications (PTMs) and glycosylation structures. Middle-down and intact CZE separations were performed in an acidified methanol-water background electrolyte on a capillary with a positively charged coating (M7C4I) coupled to an Orbitrap mass spectrometer using a commercial sheathless interface (CESI). Middle-down analysis of the IdeS-digested mAb provided characterization of PTMs of digestion fragments. High resolution CZE enabled separation of charge variants corresponding to 2X-deamidated, 1X-deamidated, and non-deamidated forms at baseline resolution. In the course of the middle-down CZE-MS analysis, separation of glycoforms of the FC/2 fragment was accomplished due to hydrodynamic volume differences. Several identified PTMs were confirmed by CZE-MS2. Incorporation of TCEP-HCl reducing agent in the sample solvent resulted in successful analysis of reduced forms without the need for alkylation. CZE-MS studies on the intact mAb under denaturing conditions enabled baseline separation of the 2X-glycosylated, 1X-glycosylated, and aglycosylated populations as a result of hydrodynamic volume differences. The presence of a trace quantity of dissociated light chain was also detected in the intact protein analysis. Characterization of the mAb under native conditions verified identifications achieved via intact analysis and allowed for quantitative confirmation of proteoforms. Analysis of mAbs using CZE-MS represents a complementary approach to the more conventional liquid-chromatography – mass spectrometry-based approaches.
Keywords: Capillary zone electrophoresis, Mass spectrometry, Monoclonal antibody, Middle-down proteomics, Native mass spectrometry
1. Introduction
Monoclonal antibodies (mAbs) are a major class of protein pharmaceuticals, with currently over 30 FDA-approved, commercially available drugs in the USA and Europe and over three hundred mAb-based products in various phases of development [1]. mAbs, as well as other protein pharmaceuticals, consist of mixtures of proteoforms, which may differ in glycosylation and other post-translational modifications (PTMs), sequence truncation, or other modifications that can occur at various stages of production, purification, or storage [2]. Given that these proteoforms may result in differences in biological activity, stability, or toxicity, comprehensive characterization of the protein therapeutic is necessary for drug approval and safety.
Conventional mAb characterization has typically been conducted by “bottom-up” liquid chromatography (LC) – mass spectrometry (MS), based on the characterization of product peptides from proteolytic enzyme digestion. However, the bottom-up approach may lead to sample preparation modifications and, importantly, precludes the ability to identify individual proteoforms, often consisting of combinations of multiple modifications [3]. Thus, methods for intact and middle-down protein analysis are being developed [4].
Analysis of intact mAbs is typically performed to determine protein molecular mass, the extent of oligomerization, and additional structural information, such as the presence of free chains [5], glycosylation [6], as well as various PTM combinations [7]. On conventional Orbitrap™ instruments, MS resolution is inversely proportional to the square root of m/z, which makes baseline resolution of higher molecular weight species challenging. This is applicable to intact mAbs, which possess multiple glycosylation sites and exhibit signals in the relatively high m/z range ~(m/z 2500–7000) [8]. In addition to intact analysis, characterization via digestion of the ~150 kDa mAb into large fragments (middle-down) has also been explored [9]. Middle-down analysis of mAbs, characterized by lower molecular weight precursors, has the potential to resolve subtle differences in the m/z domain, as well as to detect and identify low-abundance PTMs. Moreover, precursor ion selection with an analytical quadrupole on a commercial Q Exactive™ Orbitrap mass spectrometer is confined to an upper m/z limit of 2500 Th [10], thereby precluding the ability to select the most abundant charge states of an intact mAb for MS2 analysis. Conversely, limited mAb digestion (e.g., using IdeS enzyme) and ionization under denaturing conditions yield both the precursor and fragment ion species, which are amenable to analysis on a commercial Orbitrap™ MS [11]. In this study, both the middle-down and intact analyses have been explored.
Critical to the success of MS analysis of a heterogeneous mixture of a mAb is separation prior to MS detection. Typically, various 1D or 2D liquid chromatography (LC) approaches are employed either online or offline to MS. These approaches include reversed-phase liquid chromatography (RPLC) [11], size exclusion chromatography (SEC) [12], ion exchange chromatography (IEX) [13], hydrophobic interaction chromatography (HILIC) [14], or hydrophilic interaction liquid chromatography [15]. An alternative high resolution technique is capillary zone electrophoresis (CZE), which has been demonstrated to be a high efficiency separation method for the analysis of proteins [16]. In contrast to LC, CZE is based on an orthogonal underlying mechanism of separation, which relies on differences in net charge and hydrodynamic volume and does not involve the interaction of the analyte (and its surface moieties as in the case of large molecules) with the stationary phase. Therefore, CZE has the potential to provide complementary structural information to LC.
A critical component of coupling CZE to ESI-MS is the interface junction. The most straightforward CZE-ESI-MS junction involves a sheath-liquid, and there have been successful reports of protein analysis using this approach in both microcapillary and microchip formats [17–19]. An alternative approach is the use of a sheathless junction, in which the background electrolyte (BGE) is electrosprayed directly from the separation capillary without dilution from the sheath liquid [20]. A commercial sheathless system (CESI) has been introduced and shown to be highly sensitive for the analysis of peptides and proteins, including glycoproteins [6]. A recent study from our laboratory using the CESI interface has demonstrated the separation of recombinant human interferon beta-1 (rhINF-β1, ~23 000 Da), where 138 proteoforms were detected and 55 quantitated [21]. In another study, mixtures of native proteins and protein complexes, a monoclonal antibody, and the protein extract from the E. coli ribosomes were characterized using the CESI interface in one of the first examples of native CZE coupled online to ESI-MS [22].
In the present study, middle-down and intact CZE-MS have been explored for the analysis of a therapeutic mAb of the immunoglobulin type 1 (referred to as “mAb” throughout this manuscript). Experiments were performed with a positively-charged capillary coating (1-(4-iodobutyl) 4-aza-1-azoniabicyclo[2,2,2] octane iodide (M7C4I) [23]) using an acidic methanol/water background electrolyte (BGE). The middle-down analysis was performed via digestion with IdeS (“FabRICATOR”; Genovis, Lund, Sweden) protease with and without subsequent reduction. Major PTMs of the mAb, including glycosylation, oxidation, deamidation, pyroglutamation, carboxymethylation of lysine, and glycation have been observed. Confirmation of many proteoform identities was achieved by middle-down CZE-MS2 using higher collision energy dissociation (HCD). Separations of charge variants arising from 2X-deamidated, 1X-deamidated, and non-deamidated species, as well as other proteoforms differing in hydrodynamic volume, were attained. With respect to the intact mAb, we have also achieved separations of a variety of glycosylated structures, including 2X-glycosylated, 1X-glycosylated, and fully aglycosylated populations. In addition, trace quantities of the non-covalently bound light chain were observed under the denaturing conditions of the BGE. Different forms of the intact mAb observed under denaturing conditions (M7C4I capillary) were also quantitated and verified in a separate analysis performed under native conditions (polyacrylamide-coated capillary). These results demonstrate the potential of CZE-MS as a complementary approach to LC-MS in the middle-down and intact characterization of mAbs.
2. Materials and methods
2.1. Materials and chemicals
Methanol, isopropanol, acetic acid, formic acid, sodium hydroxide, sodium tetraborate, boric acid, sodium chloride, and tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl) were obtained from Sigma Aldrich (St. Louis, MO, USA). Purified mAb stocks (51.1 mg/mL, pi 8.79) were used as samples. The (M7C4I) reagent was synthesized according to previous reports [24]. IdeS was purchased from Genovis (Lund, Sweden). Micro Bio-Spin 6 columns, used for sample clean-up, were purchased from BioRad (Hercules, CA). All sheathless bare fused silica (BFS) and polyacrylamide-coated capillaries were provided by SCIEX (Brea, CA).
2.2. Sample preparation
For middle-down CZE-MS with IdeS digestion, the mAb in formulation was first diluted to 1 mg/mL in 50 mM ammonium acetate, pH 6.5. Next, IdeS was added per the manufacturer’s specifications and allowed to digest the protein for 30 min at 37°C, followed by clean-up with Micro Bio-Spin 6 columns (Bio-Rad, Hercules, CA). For subsequent reduction, TCEP-HCl was added to the cleaned-up sample to a final concentration of 5 mM, and reduction was then performed for 60 mins at room temperature. For intact CZE-MS, the mAb was diluted to 1 mg/mL in 50 mM ammonium acetate, pH 4.0, cleaned-up with Micro Bio-Spin 6 columns, immediately followed by sample injection. For native CZE-MS, the mAb was diluted to 1 mg/mL in 40 mM ammonium acetate, pH 7.5 from formulation. The mAb was then cleaned up with Micro Bio-Spin 6 columns, immediately followed by sample injection.
2.3. CZE-methods
A detailed procedure for the administration of the M7C4I reagent onto standard bare fused silica capillaries may be found in the Supporting Information section.
All CZE methods involving the M7C4I-coated capillary employed 20 kV in reverse polarity with ramp times of 1 min. Sample injections were performed at 1 psi for 10 s, corresponding to a 1.7 nL injection volume. Following each separation, a 5-min ramp down time to 1 kV was programmed to protect the integrity of the porous sprayer tip.
For middle-down CZE-MS/CZE-MS2 experiments with a BGE of 50% methanol, 1% formic acid, the optimum spray potential was determined to be 1.3 kV and was employed to maintain stable nanoESI. For intact CZE-MS experiments involving the 10% isopropanol, 0.2% formic acid BGE, a 1.4 kV spray potential was used.
For native CZE-MS analysis of the mAb, a polyacrylamide-coated capillary (SCIEX, Brea, CA) was used. Prior to each injection, a series of rinses of the separation and conductive lines were first performed. For the separation capillary these included: 0.1 M hydrochloric acid (100 psi, 5 min), DDI water (100 psi, 5 min), followed by the 40 mM ammonium acetate, pH 7.5 BGE (100 psi, 10 min); the conductive capillary was rinsed with 10% acetic acid at 75 psi for 4 min. Sample injection was performed at 2.5 psi for 15 s, corresponding to approximately 6 nL of the sample (0.8% of the capillary volume). Separations were performed for 30 min at 20 kV (in normal polarity) with a ramp time of one minute. Due to the significant suppression of EOF on polyacrylamide-coated capillaries, a supplemental pressure of 3 psi was applied to generate a flow of the solvent and facilitate the migration of analytes towards the MS detector. At the end of the run, a ramp-down from 20 to 1 kV over 5 min was performed back to the start conditions for the next injection.
2.4. MS instrumentation
In our experiments, a Q Exactive™ Plus Orbitrap™ MS (Thermo Fisher Scientific, Bremen, Germany) was employed in both the intact protein and standard modes of operation. For both middle-down and intact CZE-MS, a 50 eV potential was established to provide for the optimum insource collision-induced dissociation energy (in-source CID), allowing for enhanced desolvation of the precursor ions. For middle-down schemes on the Q Exactive™ Plus, the analysis was performed with an automatic gain control (AGC) of 106, a maximum injection time of 250 ms, a resolving power of either 17 500 or 140 000 at m/z 200, an inlet transfer capillary temperature of 110°C, and an averaging over three microscans. The nominal resolving power of 17 500 was used in CZE-MS experiments, while 140 000 was selected for precursor ion scans in CZE-MS2 experiments. In CZE-MS experiments with intact forms, detection settings were identical to those described above, using a nominal MS resolving power of 17 500.
In CZE-MS2 experiments, a “Top 10” method was used. Precursor scans were performed as described above. For MS2 scans, instrument resolving power was set to 140 000 at m/z 200 with averaging over five microscans. AGC was set to 5 × 105, with a maximum injection time of 250 ms. An isolation window of 6 Th was selected, and a 23 eV potential was determined to provide for the optimum normalized collision energy.
In native CZE-MS analysis of the mAb, a polyacrylamide-coated capillary was coupled to the Q Exactive™ Plus Orbitrap MS. A 1.4 kV spray potential was determined to be the optimum ESI voltage. Protein collisional cooling and trapping were performed in the HCD cell of the instrument, as opposed to the C-trap in the intact (denaturing) and middle-down experiments. Given the relatively high m/z range for an intact mAb under native conditions (5000–7000 m/z), detection required optimization of the in-source CID voltage and corresponding voltages in the HCD cell. In the native analysis, a potential of 40 eV for in-source CID, in addition to a voltage of 75 eV in the HCD cell were used for optimum collisional cooling and desolvation. Trapping and collisional cooling were also performed at elevated gas pressures in the HCD cell, corresponding to approximately 1.02e-4 mbar in native CZE-MS experiments. This trapping pressure is considerably higher than standard trapping pressure used in intact (denaturing) (4.37e-5 mbar) and reduced trapping pressure (1.80e-5 mbar) used for middle-down analyses. In native CZE-MS, an AGC of 2.0e5 and a maximum injection time of 50 ms were used. Finally, the instrument was scanned from 1500–10 000 m/z at a resolving power of 17 500 at 200 m/z.
2.5. Data analysis
Deconvolution of CZE-MS experiments was achieved with BioPharma Finder 1.0 (Thermo Fisher Scientific). Analyses of CZE-MS2 and native CZE-MS data were performed with Byonic (v. 2.5.6) and Intact Mass (v. 1.6) software tools (Protein Metrics, San Carlos, CA), respectively.
For deconvolution of CZE-MS experiments performed at 17 500 resolving power, the Sliding Window algorithm with ReSpect was used. For isotopically resolved data acquired using a resolving power of 140 000, the Sliding Window algorithm using Xtract was employed.
For analysis of CZE-MS2 experiments acquired on the Q Exactive™ Plus, Byonic (version 2.5.6) (Protein Metrics, San Carlos, CA) was utilized. Searches were performed using fully specific digestion with a maximum of two missed cleavages. A 10 ppm precursor mass tolerance and a 20 ppm fragment mass tolerance were used in all searches, along with a 1% false discovery rate (FDR). Common and expected PTMs were manually added to the search bar and used as dynamic modifications. Modifications included oxidation, pyroglutamation, deamidation, N-terminal methionine-loss, glycation, di-oxidation, and carboxymethylation of lysine. A database of 38 common biantennary glycans was also included in the searches.
For deconvolution of native CZE-MS experiments using Intact Mass software, a deconvolution range corresponding to 5000–6600 Th was selected with a 40 ppm tolerance. A range corresponding to 17–20 min of the electropherogram was selected for deconvolution. The above deconvolution range was selected to allow for accurate deconvolution and quantitation of 2X-glycosylated, 1X-glycosylated, and aglycosylted populations of the mAb.
3. Results and discussion
3.1. Middle-down analysis
3.1.1. Products of proteolytic digestion and reduction
In this work, middle-down analysis of the mAb involving digestion with IdeS was explored. Supporting Information Fig. 1 shows the expected products from this digestion. Briefly, digestion with IdeS resulted in the cleavage between Gly residues in the hinge region of the antibody, yielding F(ab’)2 (98.4 kDa) and two FC/2 (~24 kDa) fragments. Subsequent reduction resulted in the dissociation of F(ab’)2, yielding two light chains (Lc) (23.2 kDa) and two FD fragments (26.0 kDa), in addition to the two FC/2 fragments.
3.1.2. CZE-MS separation parameters
Despite the recent success in the characterization of rhINF-β1 using a positively charged cross-linked polyethyleneimine (cPEI) coating [21], intact and middle-down analyses of the mAb under the same conditions yielded limited proteoform separations. On the cPEI-coated capillary under acidic conditions, a higher electroosmotic flow (EOF) mobility relative to the electrophoretic (free solution) mobilities of the intact and middle-down products of the mAb was observed. Typically, the EOF mobility was two- to threefold higher than analyte electrophoretic (free solution) mobilities, precluding separation of the mAb proteoforms (results not shown). Hence, we aimed at increasing separation performance (resolution, Rs) by reducing the EOF (see Supporting Information Eq.1). A positively charged capillary coating with a lower density of charged groups relative to cPEI was thus selected (M7C4I) [23]. In comparison to cPEI, the M7C4I-coated capillary has been reported to generate a lower reversed EOF [25].
We then explored a variety of BGEs under acidic conditions to optimize separation on the M7C4I-coated capillary. Supporting Information Fig. 2 shows a comparison of different BGEs for separation of IdeS-digested, reduced mAb. For the digestion products Lc, FC/2, and FD, among the evaluated conditions, the BGE yielding the lowest EOF mobility and highest solute apparent mobilities was found to be 50% methanol, 1% formic acid (FA), which was then used in the further experiments. Additional details are provided in the Supporting Information section. Supporting Information Table 1 provides a summary of repeatability experiments performed on the M7C4I-coated capillary with several different samples, using different capillaries, and on different days. In agreement with previous reports [26], reproducibility on the M7C4I-coated capillary was found to be in the range of 2–3% CV.
The relative migration order of the mAb digestion fragments proceeded with Lc (23.2 kDa), followed by FC/2 (~25.2 kDa), and then FD (~26 kDa). If only differences in the hydrodynamic volume are considered, and the hydrodynamic volume is assumed to be directly proportional to analyte’s molecular mass, the relative migration order of the digestion fragments is expected to be inverted on a positively-charged capillary operated in reverse polarity (see Supporting Information section). However, since the migration time is proportional to analyte’s charge, species exhibiting the lowest positive charge are expected to migrate fastest. Thus, it is inferred that the Lc fragment possessed the lowest positive charge in solution, resulting in the fastest migration time, followed by FC/2 and then FD. These relationships are summarized in Supporting Information Eqs. 2 and 3.
3.1.3. Analysis of IdeS-digested, nonreduced mAb
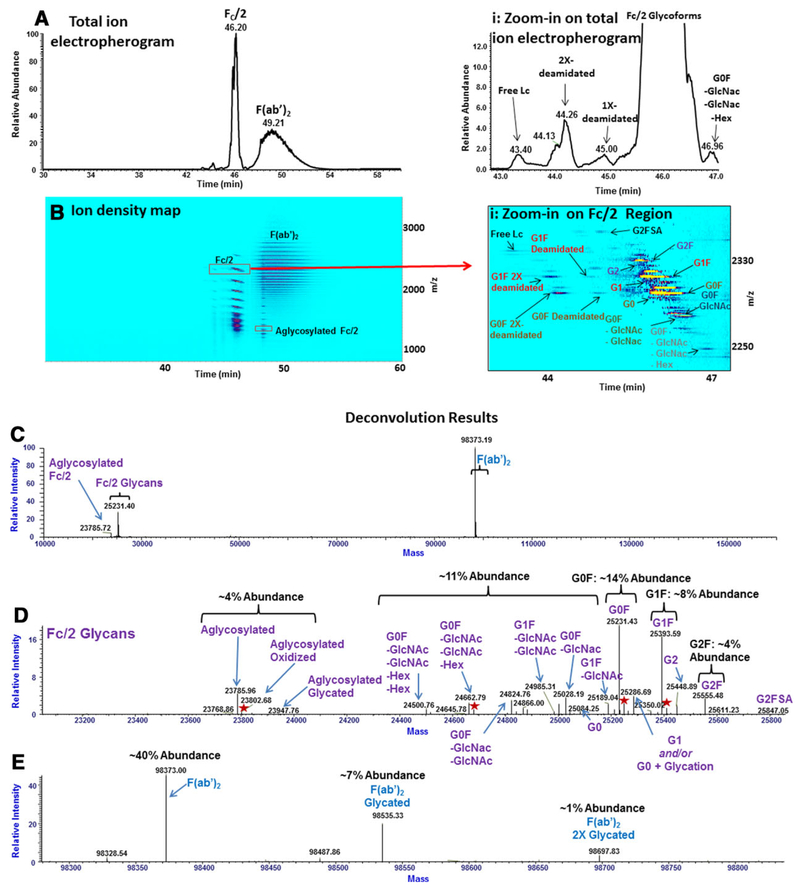
IdeS-digested mAb was analyzed on the M7C4I coated-capillary with a 50% methanol, 1% FA BGE, as shown in Fig. 1. Separation of the major digestion fragments, FC/2 and F(ab’)2, was achieved at baseline resolution (Figs. 1A and B). If FC/2 and F(ab’)2 digestion fragments differed in the hydrodynamic volume alone, and the hydrodynamic volume is assumed to be proportional to the molecular mass, migration of the F(ab’)2 fragment is expected to precede the migration of FC/2 on a positively-charged capillary operated in reverse polarity. However, as the charge is considered, the comparatively higher positive charge of the F(ab’)2 fragment in solution resulted in the slower migration of this fragment than the FC/2. Deconvolution of the CZE-MS electropherogram resulted in the identification of multiple forms for these two digested fragments with approximate relative abundances indicated in Fig. 1C–E. For the ~24 kDa FC/2 fragment, 15 different glycoforms, and an aglycosylated form were identified. Sequential additions of two deamidations were detected for G0F and G1F glycoforms, in addition to a single deamidation for the G2F glycoform. For several glycoforms of FC/2 and aglycosylated FC/2 species, low-level oxidations, as well as oxidized and glycated forms were identified (Fig. 1D).
Figure 1.
A–E. Analysis of the IdeS-digested mAb. (A) The total ion electropherogram. (A(i)) 2X- and 1X-deamidated FC/2 forms are resolved from the non-deamidated species and are shown as an inset to the total ion electropherogram. (B) The ion density map matching the electropherogram in (A). Inset (B(i)) shows a zoomed-in view on the map in (B) representing the 11+ charge state of the FC/2 species. (C) Deconvoluted mass spectrum corresponding to the FC/2 and F(ab’)2 fragments. (D,E) Insets into the regions corresponding to FC/2 and F(ab’)2 fragments, respectively, in (C). Various oxidized proteoforms in (D) are labeled as red stars. Approximate abundances of FC/2 and F(ab’)2 proteoforms are shown.
For the IdeS-digested mAb experiment, relative ratios of a variety of proteoforms are reported in Figs. 1D and E. As an example, the G0F glycoform was found to be the most abundant FC/2 species at roughly 14%. The G1F was the second-most abundant glycoform, at approximately 8%, followed by the G2F glycoform (4%). Proteoforms of the aglycosylated FC/2 were also present at approximately 4% (Fig. 1D). Supporting Information Table 2 shows relative abundances of several proteoforms and different populations of the Ides-digested mAb between three different experimental replicates. Quantitative measurement errors were found to be within 3% RSD for the non-reduced G0F and G1F glycoforms of the FC/2 fragment. In the context of glycoform analysis, one consideration involves the release of sialic acids from glycoproteins following exposure to acidic conditions and organic solvents [27]. Conceivably, the sample preparation and analytical conditions utilized in these experiments may not be suitable for a glycoprotein possessing a high content of sialic acid residues. However, in the case of the mAb under investigation in these experiments, while a comprehensive characterization of released glycans was not performed, analysis of the intact mAb under milder denaturing conditions and native conditions (see the ‘Intact CZE-MS analysis of the mAb” section and Supporting Information Fig. 11), corroborated that the majority of identified glycoforms of the mAb were composed of neutral glycans devoid of sialic acids.
As shown in the ion density map in Fig. 1B(i), partial separations of individual glycoforms of the FC/2 fragment were achieved. FC/2 proteoforms possessing larger glycans (e.g., G2F) are characterized by larger Stokes radii (R) than those with smaller glycans (e.g., G0F) and the aglycosylated FC/2, and thus also possess lower electrophoretic mobilities, larger apparent mobilities and faster migration times (see Supporting Information Eqs. 2 and 3). As the majority of identified FC/2 proteoforms contained neutral, biantennary glycans, their migration order is expected to be governed mainly by variations in the hydrodynamic volumes.
As shown in the inset in the total ion electropherogram in Fig. 1A(i) and in the ion density map in Fig. 1B(i), baseline CZE resolution was achieved between 2X-deamidated, 1X-deamidated, and non-deamidated glycoforms of FC/2. Each deamidation event in proteins results in an increase in the molecular weight of each proteoform by 0.96 Da and a discrete reduction of charge of the molecule by one, as an asparagine residue is converted to a mixture of aspartic and iso-aspartic acids (while less common, deamidations of glutamine and the protein N-terminus result in the same delta mass shift) [28]. Since a 0.96 Da molecular mass increase does not yield a detectable increase in the hydrodynamic volume, separations of the deamidated forms were likely to be effected by differences in the net charge. In CZE, electrophoretic mobility (μEP) is proportional to the analyte charge (q) (see Supporting Information Eq. 3). Thus, species imparted by deamidation and possessing lower net positive charge migrate faster than their non-deamidated counterparts on a positively-charged capillary operated in reverse polarity. The experimentally-observed migration order for the glycoforms of FC/2 was found to be in accord with their net charges, revealing shorter migration times of the least positively-charged 2X-deamidated species, followed by the 1X-deamidated and then by the non-deamidated glycoforms. Like deamidation, baseline separation of the G2FSA glycoform from the G2F glycoform was also achieved as a result of the additional negative charge imparted by the presence of a sialic acid residue (Fig. 1B(i)).
Figure 1D shows that several oxidized FC/2 glycoforms, as well as an oxidized and a glycated form of the aglycosylated FC/2, were identified by deconvolution. In the case of oxidation, no charge variation is expected. Similarly, while glycation results in a minor increase of the protein acidity (~0.05–0.09 pI units [29,30]), under the employed acidic CZE-MS conditions, no charge variation is expected to be detectable, as the Amadori product maintains the same charge and a fully protonated status as the non-glycated variant [28]. With these modifications, CZE separations were impeded by the lack of a significant charge variation under the acidic conditions. Differences in hydrodynamic volumes, represented by a +16 Da change for oxidized forms and +162 Da change for glycated forms, were found to be insufficient to enable CZE separation based on subtle differences in hydrodynamic volumes of proteoforms.
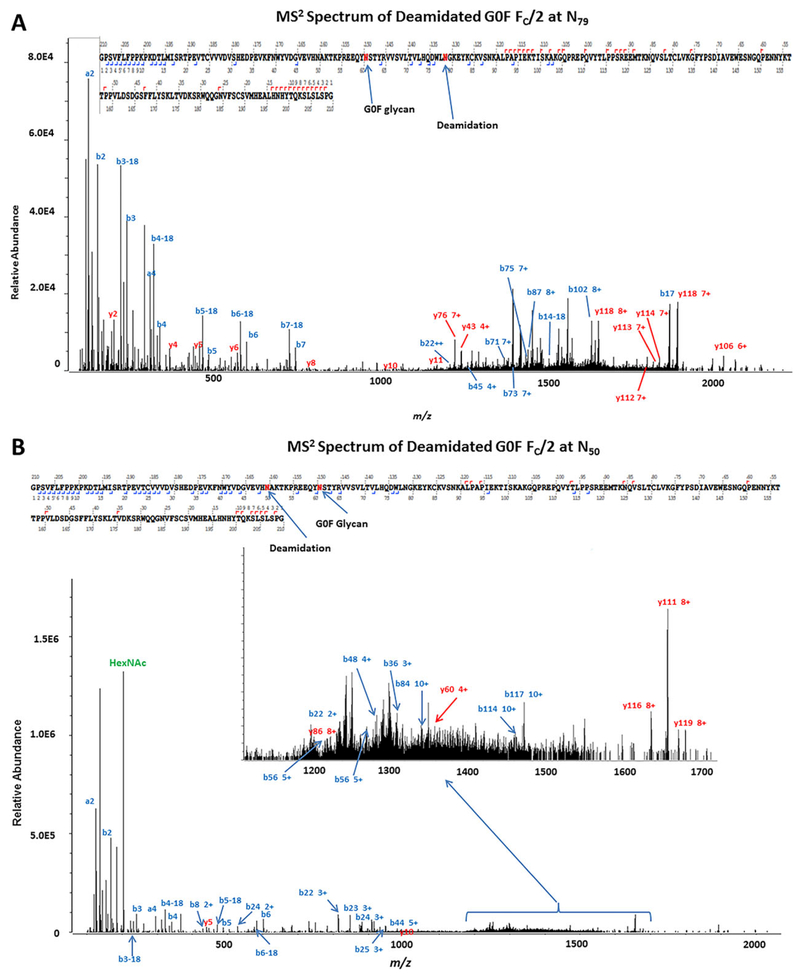
A CZE-MS2 “Top N” method was then used to confirm and determine the localization of PTMs attached to specific amino acid residues. For example, for the G0F FC/2 fragment, deamidations at residues N79 (Fig. 2A) and N50 (Fig. 2B) were detected. 1X-deamidation assigned previously to the G1F FC/2 protein by accurate mass was also confirmed by CZE-MS2. CZE-MS2 enabled confirmation of most biantennary glycans ascribed to the FC/2 fragment from previous CZE-MS experiments. CZE-MS2 also resulted in the identification of a glycated form (at residue K90) of the G0F FC/2 fragment (Figure S3). Analysis of CZE-MS data resulted in the identification of a G0 FC/2 form, in addition to a G1 FC/2 proteoform. However, identification of a glycated form of G0 FC/2 by CZE-MS2 suggests that the peak corresponding to the G1 FC/2 proteoform could in part be a combination of G1 FC/2 and glycated G0 FC/2, represented by the same delta mass shift (see Fig. 1D).
Figure 2.
A and B. Tandem mass spectra of proteoforms of the FC/2 fragment from the IdeS-digested mAb experiment. (A) CZE-MS2 HCD mass spectrum of the deamidated G0F FC/2 fragment. Glycosylation and deamidation were confidently identified at residues N60 and N79, respectively. (B) Confirmation of deamidation at residue N50 of the G0F FC/2 proteoform by CZE-MS2. The MS2 spectrum shows the identified a-, b-, and y-fragment ions. The inset in (B) shows a portion of the mass spectrum representing fragment ions used to identify deamidation. The top panels in (A) and (B) show sequence coverage obtained by CZE-MS2.
Turning to the 98.4 kDa F(ab’)2, two glycations were found after deconvolution of the electropherogram. Unmodified F(ab’)2 was detected at 40%, while 1X-glycated and 2X-glycated F(ab’)2 were present at roughly 7 and 1%, respectively (Fig. 1E). However, the relatively large size of the F(ab’)2 precluded identification of other PTMs by MS, as small differences in m/z were unresolved by the mass spectrometer. Moreover, such forms exceeded the 2500 Th limit of precursor selection by the analytical quadrupole of the mass spectrometer used in these experiments. Thus, characterization of proteoforms of F(ab’)2 required a further decrease of the 98.4 kDa F(ab’)2 protein into smaller fragments by means of reduction.
3.1.4. Analysis of IdeS-digested, reduced mAb
Reduction of IdeS-digestion products, FC/2 and F(ab’)2, decreased the molecular weights of species, allowing for improved characterization of the mAb. The formed products included Lc (23.2 kDa) and FD (26.0 kDa), along with the reduced FC/2 fragment (~24.0 kDa). However, reduction of IdeS-digestion products with TCEP-HCl resulted in the introduction of significant artificial heterogeneity (i.e., multiple isoforms of scrambled disulfides for each digestion fragment) if the reducing agent was removed from the sample buffer prior to injection into the capillary, as shown in Supporting Information Fig. 4A. Conversely, the inclusion of TCEP-HCl eliminated scrambled disulfide isoforms of IdeS-digestion products and resulted in a simplification of the electropherogram (Supporting Information Fig. 4B). The use of TCEP-HCl also simplified sample preparation, as an alkylation step was not required. For the middle-down schemes described below, limited proteolytic digestions were followed by reduction with TCEP-HCl. The reduced samples were then buffer-exchanged into water containing up to 5 mM TCEP-HCl.
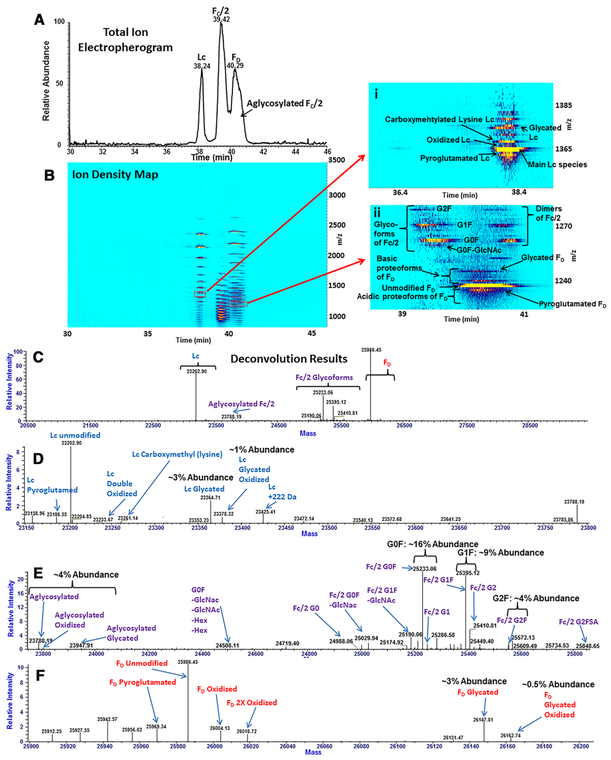
Figure 3 shows the characterization of the IdeS-digested, reduced mAb with TCEP-HCl included in the sample buffer during injection. While the analysis of the IdeS-digested mAb facilitated the characterization of proteoforms of the FC/2 fragment, reduction following digestion enabled a more comprehensive analysis of the F(ab’)2 fragment. The latter enabled characterization of proteoforms of the formed Lc and FD fragments (Figs. 3A and B). Shown in Figs. 3C–F, the deconvolution of the entire electropherogram resulted in the identification of numerous proteoforms of Lc, FD, and FC/2 digestion fragments.
Figure 3.
A-F Analysis of the IdeS-digested, reduced mAb. (A) Total ion electropherogram of digestion fragments. (B) The corresponding ion density map with zoomed-in regions for Lc (B(i)) and FD (B(ii)), shown as separate insets. (C) Deconvoluted spectrum, with zoomed-in views of regions corresponding to Lc (D), FC/2 (E), and FD (F).
For the Lc fragment shown in Fig. 3D, proteoforms possessing pyroglutamation, oxidation, carboxymethylation of lysine, and glycation were observed. Depicted in Fig. 3B(i), the ion density map of the Lc 17+ charge state shows the resolution of the individual species in both CZE and MS domains. In the case of glycation, as described previously, no charge variation between glycated and unmodified forms of Lc was observed. Like the aglycosylated FC/2 species, glycation of the Lc fragment did not result in a sufficient hydrodynamic volume change to enable separation from unmodified Lc. Thus, glycated Lc co-migrated with unmodified Lc. Carboxymethylation of lysine is an advanced glycation end-product characterized by a +58 Da delta mass shift [31] with no accompanying change in charge under the acidic conditions, similar to glycation. Shown in Fig. 3B(i), the carboxymethylated lysine form of Lc was unresolved from the unmodified variant. Like glycation, the variation in hydrodynamic volume due to carboxymethylation of lysine was found to be insufficient to enable CZE separation under the selected conditions. For the same reason, double-oxidized forms of the Lc, as well as its glycated and oxidized proteoforms, were unresolved from the unmodified variant. The pyroglutamated Lc form also co-migrated with unmodified Lc, as no charge variation occurred during the conversion from N-terminal glutamate to pyroglutamic acid.
Similar to the IdeS-digested, non-reduced middle-down analysis of the mAb (see Fig. 1D), a number of glycosylated FC/2 proteoforms, aglycosylated FC/2, oxidized aglycosylated FC/2, and glycated aglycosylated FC/2 species were observed for the IdeS-digested, reduced mAb. Figure 3E shows all the identified proteoforms of the reduced FC/2 fragment. FC/2 glycoforms were identified in approximate abundances of 4% for aglycosylated FC/2 species, 16% for G0F FC/2, 9% for G1F FC/2, and 4% for G2F FC/2. Partial separation of numerous glycoforms of the FC/2 fragment is demonstrated in Fig. 3B(ii), which shows the ion density map for the IdeS-digested, reduced mAb experiment. Similar to the IdeS-digested mAb experiment, FC/2 proteoforms characterized by larger glycans (e.g., G2F) migrated faster due to the hydrodynamic volume effect. Separations of 2X-deamidated from 1X-deamidated, from non-deamidated FC/2 glycoforms, were achieved as a result of differences in a single charge imparted by each deamidation. The least positively-charged, 2X-deamidated analytes migrated first, followed by the 1X-deamidated and then by the unmodified FC/2 glycoforms.
Additionally, analysis of the IdeS-digested, reduced mAb resulted in the identification of multiple proteoforms of the FD fragment. Specifically, pyroglutamated, oxidized, double-oxidized, glycated, and combined glycated and oxidized FD forms were detected by deconvolution, as shown in the deconvoluted mass spectrum in Fig. 3F.
From deconvolution results of the IdeS-digested mAb analysis, approximately 7% of the F(ab’)2 was found to be glycated, along with an approximate 1% relative abundance for the 2X-glycosylated species (Fig. 1D and E). These results were corroborated by the IdeS-digested, reduced mAb experiment, which revealed approximately half of the glycation attributed to the FD (Fig. 3D) and half to the Lc fragments (Fig. 3F).
CZE-MS2 analysis of the IdeS-digested, reduced mAb was performed to validate identifications of PTMs of major digestion fragments and to establish localization of the modified residue sites. A key limitation of the IdeS-digested mAb experiment was the inability to analyze the large F(ab’)2 protein by CZE-MS2. Reduction of IdeS-digestion products of the mAb facilitated the analysis of FD and Lc fragments, as charge-state distributions of precursor ions were below the 2500 Th threshold for selection for CZE-MS2 analysis. From the CZE-MS2 analysis, with high confidence, two different oxidations were identified on the Lc fragment at R24 (Supporting Information Fig. 5) and R108 (Supporting Information Fig. 6) residues. These assignments were corroborated by the CZE-MS deconvolution results illustrated in Fig. 3D. Also, N-terminal pyroglutamation for the FD fragment was also confirmed by the CZE-MS2 analysis (Supporting Information Fig. 7).
A comparison of CZE separations of IdeS-digested mAb and IdeS-digested, reduced mAb revealed a difference in migration times of both the glycosylated and aglycosylated FC/2 fragments. For example, in the case of the glycosylated FC/2 proteoforms from the IdeS-digested sample, migration time was approximately 46.2 min (Fig. 1A-B). However, for the IdeS-digested, reduced sample, the migration time of the identical forms was approximately 39.5 min (Figs. 3A-B). This observation may be explained by different degrees of unfolding as a result of disulfide bond cleavage. Two phenomena are observed simultaneously. On the one hand, reduction results in protein unfolding and greater exposure of internal residues to the outside environment. This is evident, in part, by different charge state distributions on ESI mass spectra detected for identical forms between IdeS-digestions of the mAb with and without reduction. Supporting Information Fig. 8 shows a comparison of ESI mass spectra of IdeS-non-reduced glycosylated FC/2 and IdeS-reduced glycosylated FC/2 fragments. In the case of the non-reduced glycosylated FC/2 species, the detected charge state distribution was constrained between [M+11H]11+ and [M+19H]19+ (highest abundance: [M+11H]11+) species. For the reduced glycosylated FC/2 species, the distribution shifted to [M+14H]14+ and [M+28H]28+ charge states (highest abundance: [M+25H]25+). While these charge state distributions may not be correlated directly to analyte charge in solution, a subtle increase in charge is nevertheless expected as a result of unfolding. On the other hand, protein unfolding from reduction is expected to result in a significant increase in the hydrodynamic volume. In CZE with a positively-charged capillary coating, species with larger hydrodynamic volumes experience faster migration times (see Supporting Information Eqs. 2 and 3). This trend is confirmed by our experiments with the reduced glycosylated FC/2 proteoforms. Due to partial unfolding, the latter are characterized by increased hydrodynamic volumes and were found to migrate faster than the non-reduced variants. A similar trend was also observed for the aglycosylated FC/2 species. These effects highlight the sensitivity of CZE to differences in analyte hydrodynamic volume.
On a conventional mAb of the IgG1 type, the FC fragment is characterized by two intra-disulfide bonds, one in the CH2 domain and one in the CH3 domain [32]. Based on a previous report, there is a buried, highly-conserved disulfide bond within the CH3 domain that connects two β-sheets. Proper assembly of a generic IgG1 mAb is dependent in part on dimerization of the CH3 domains from both FC/2 fragments [33]. However, several studies have suggested that the presence of the intra-disulfide bond in the CH3 domains is not a requirement for their dimerization [33–35]. Moreover, it was determined that the secondary and tertiary structures of reduced and oxidized CH3 domains are similar, albeit some differences were nevertheless observed [33]. The biotherapeutic mAb reported in this study, as common for IgG type 1 antibodies, contains a single intra-disulfide bond in each of the CH2 and CH3 domains (determined by bottom-up proteomic analysis of the mAb, not reported in this manuscript). Thus, oxidized and reduced FC/2 fragments are expected to possess a +4 Da difference in molecular masses, stemming from reduction of the two internal disulfide bonds. However, comparison of molecular masses of observed reduced FC/2 fragments and their non-reduced counterparts, arising from IdeS-digestion alone, reveal a difference of only +2 Da (for example, 25,231.43 Da for the reduced FC/2 G0F proteoform vs. 25,233.06 Da for the non-reduced form, or 23,785.96 Da for the aglycosylated FC/2 vs. 23,788.19 Da for the non-reduced form), presumably corresponding to a single reduced internal disulfide bond. Since the disulfide present within the CH3 domain is reportedly buried within the domain [33], we hypothesize that this disulfide may have remained in the oxidized state, which resulted in +2 Da difference observed between the reduced and non-reduced FC/2 proteoforms. Presumably, reduction of the disulfide bond in the CH3 domain may not have occurred as TCEP-HCl was added to an IdeS-digested mAb in an ammonium acetate buffer at physiological pH, where digested fragments FC/2 and F(ab’)2 were not denatured. Even though the conformational differences between the reduced and non-reduced forms of the CH3 domain may not be very significant [33], in our study, differences in migration times of reduced and non-reduced FC/2 fragments are likely explained by the hydrodynamic volume change imparted by the reduction of the CH2 domain of the FC/2 fragment, where the internal disulfide bond is comparatively more exposed to the molecule’s environment than the one in the CH3 domain.
3.2. Intact CZE-MS characterization of the mAb
In the middle-down analysis, characterization of FC/2 fragments revealed individual glycosylation states. However, an intact mAb may be composed of asymmetric glycans across its two FC/2 domains. This information is not accessible by most middle-down analysis approaches involving reduction. Consequently, the intact mAb analysis is an important complementary approach to middle-down experiments allowing for the characterization of glycosylated structures of the mAb.
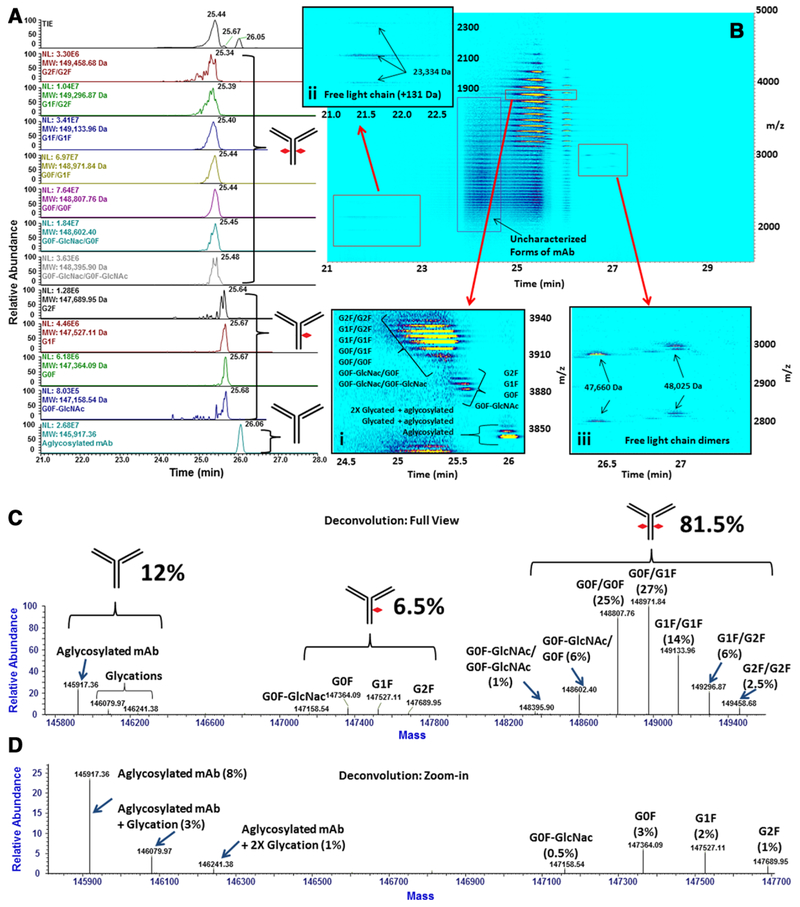
Initial analysis of the intact mAb in the 50% methanol, 1% FA BGE used in the middle-down experiments resulted in the poor separation of proteoforms of the intact mAb, as shown in Supporting Information Fig. 9. Figure 4 shows the results of CZE-MS analysis of the intact mAb under the optimized BGE conditions, composed of 10% isopropanol, 0.2% FA. Our experiments of the intact protein have revealed the complete glycosylation profile of the mAb, in addition to the elucidation of a trace quantity of Lc bound non-covalently to the remainder of the molecule. Figure 4A shows the extracted ion electropherograms of major glycosylated proteoforms of the mAb, which are accompanied by an ion density map, shown in Fig. 4B. Figure 4B(i) shows the 38+ charge state of the intact molecule, revealing all three detected populations of the mAb in both CZE and MS domains. Figs. 4B(ii)-(iii) show species stemming from dissociation of light chains of the mAb. Species identified in Fig. 4B(ii) correspond to a dissociated Lc with an uncharacterized +131 Da delta mass shift relative to unmodified Lc. Species shown in Fig. 4B(iii) were presumed to be dimers of the Lc.
Figure 4.
A–D. Denaturing analysis of the intact mAb. (A) The total ion electropherogram (TIE), along with extracted ion electropherograms (EIEs) of major glycoforms of the mAb. (B) The ion density map for the CZE-MS experiment shown in (A). The inset (B(i)) shows the ion density map of the 38+ charge state of the mAb. CZE-MS signal intensities of glycoforms, such as 2X-glycosylated, 1X-glycosylated, and aglycosylated are illustrated. Insets B(ii)) and B(iii)) show the detected free chains of the mAb. (C) Deconvolution results showing major glycosylation states of the intact mAb for the electropherogram region between 24.7 min and 26.0 min. Quantitative ratios of each population of the mAb (2X-glycosylated, 1X-glycosylated, aglycosylated) are specified. (D) A zoomed-in view of the deconvolution spectrum to ~25% relative abundance shows the detected aglycosylated and 1X-glycosylated proteoforms of the mAb.
The deconvoluted mass spectra in Fig. 4C and D revealed the presence of three types of populations of mAb glycans. Namely, (i) the 2X-glycosylated mAb, with glycans present on both FC/2 domains; (ii) 1X-glycosylated mAb, with glycans present on one of the two FC/2 domains; and (iii) aglycosylated mAb with no bound glycans. Figure S10 shows repeatability of quantitation results for 2X-glycosylated, 1X-glycosylated, and aglycosylated populations of the mAb across three experimental replicates. These results, combined with obtained ratios shown in Fig. 4C, resulted in approximate ratios of the 2X-glycosylated mAb to the 1X-glycosylated, to the aglycosylated of 84.3 ± 2.49% to 4.63 ± 28.4% to 11.1 ± 7.68%, respectively. A summary of these results is also presented in Supporting Information Table 3. In addition to the aglycosylated form detected at 8.5 ± 6.79% relative abundance, up to two glycations were detected for the aglycosylated mAb, with relative abundances of approximately 2.25 ± 22.2% for the 1X-glycated variant and 0.75 ± 47.1% for the 2X-glycated proteoform (Figs. 4C-D). CZE separation was achieved for the mAbs with different extents of glycosylation, shown in Fig. 4A(i). As the majority of detected glycan moieties are neutral structures, it is assumed that individual populations were resolved due to differences in the hydrodynamic volume. Relative migration proceeded in the order of the 2X-glycosylated, to the 1X-glycosylated, to the aglycosylated mAb, according to a decrease in the hydrodynamic volume.
Trace quantities of the dissociated Lc and Lc dimers were also detected, with migration times of 21.5 and 26.5 min, respectively (Fig. 4B(ii)-(iii)). These species were detected at less than 1% relative abundances.
In total, four experimental replicates of the intact mAb analysis under denaturing conditions (sample solvent: 50 mM ammonium acetate, pH 4.0; BGE: 10% isopropanol, 0.2% formic acid) were performed. In the first experimental replicate (Fig. 4), a fresh sample was analyzed on a freshly-administered capillary coating. In the experimental replicates 2–4 (Supporting Information Fig. 10), the same analysis was conducted on a rejuvenated capillary coating on the following day using the same sample that was stored at 10°C between the analyses. Experimental replicates 2–4 were performed on the same day over a time course of several hours. As shown in Supporting Information Table 3, the relative abundance of the 2X-glycosylated mAb species increased approximately 5% over time (~24 hours, comparing replicates 1 and 4), considering abundancies of all detected proteoforms, while the relative abundances of the 1X-glycosylated and aglycosylated mAb populations decreased approximately 3 and 2% (comparing replicates 1 and 4), respectively (Fig. 4C, Supporting Information Fig. 10, and Supporting Information Table 3). Presumably, this may be attributed to different degrees of stability of the mAb populations in denaturing and acidic sample solvent. Since mAb glycosylation is known to improve the stability of the protein [36], the populations of the mAb possessing a lower extent of glycosylation (1X-glycosylated and aglycosylated) may exhibit a higher propensity for degradation as the mAb is buffer-exchanged from formulation, where it exhibits greater stability, to the denaturing sample solvent and incubated over a period of several hours. Thus, over time, the relative abundance of the 2X-glycosylated population of the mAb may increase among all detectable mAb proteoforms, while those of the 1X-glycosylated and aglycosylated may decrease. In Fig. 4B, a distinct electrophoretic zone between 23.7 and 24.8 min, labeled as “uncharacterized mAb” may be observed. Due to the highly heterogeneous nature of the averaged mass spectrum across this region and relatively low signal intensities of detected species, reliable deconvolution of this region could not be obtained. Conceivably, mAb proteoforms imparted by degradation from acidic and denaturing sample storage conditions may have been present within this electrophoretic region. However, further investigation is needed to validate this hypothesis.
To verify abundances of various proteoforms of the mAb determined in the intact analysis, we also characterized the intact mAb under native conditions using a polyacrylamide-coated capillary coupled to the Q Exactive Plus™ MS (Supporting Information Fig. 11). In these experiments, the performance of the M7C4I-coated capillary was suboptimal, as under native analysis conditions involving a BGE at physiological pH, analytes would possess significantly higher apparent mobilities (due to the high EOF mobility), resulting in a much lower resolution. Thus, the commercial polyacrylamide-coated capillary, possessing negligible EOF, was selected for native experiments. While the separation of individual glycosylation states (2X-glycosylated, 1X-glycosylated, and aglycosylated) was not achieved (Supporting Information Fig. 11A and B), this approach resulted in the detection of mAb dimers at less than 1% relative abundance. These species co-migrated with the main forms of the mAb (Supporting Information Fig. 11A and B). The detection of aggregates is an important advantage of the analytical technique for characterization of mAbs. A previous study out of our lab also reported on dimers detected in native CZE-MS analysis of a different mAb [22].
Similar to the experiments performed under denaturing conditions (Fig. 4), dissociated light chain, light chain dimers, and a proteoform of the intact mAb missing two light chains were detected under native analysis conditions. These forms were baseline-resolved from the main species of the mAb by CZE-MS (Supporting Information Fig. 11A and B). The total relative abundances of these structures, determined from the analysis of the intact mAb under native conditions (Supporting Information Fig. 11A and B), were identified at less than 1% and were corroborated with the results obtained from the denaturing experiments shown in Fig. 4. There are two plausible explanations for the presence of such incomplete mAb populations. The first explanation involves cellular production and secretion of an incompletely assembled mAb (a structure composed exclusively of two covalently-bound heavy chains) along with the corresponding free light chains, which are thus represented in the final mAb formulation in trace amounts in addition to the predominant species of the fully assembled mAb. The second explanation involves a mAb structure, present in trace quantities, where individual light chains are bound non-covalently to a covalent heavy chain dimer and dissociate following buffer exchange from formulation and/or exposure to non-native conditions. It is not clear whether one of the two scenarios or their combination is responsible for the presence of the partially dissociated mAb, but it should be mentioned that mAbs composed of individual chains held together exclusively via non-covalent forces occur under native conditions and have been reported previously [37].
Abundances of major forms of the mAb identified under denaturing conditions (see Fig. 4 and Supporting Information Fig. 10) were verified with the abundance values resulting from deconvolution of the main electrophoretic peak observed in the native experiment. Supporting Information Fig. 11C shows the deconvolution result for the main peak observed in Supporting Information Fig. 11A. Relative abundances of individual proteoforms of the mAb were comparable between native and denaturing experiments and across two different deconvolution algorithms. For example, the aglycosylated mAb was present at approximately 7% relative abundance under native conditions and 8% under denaturing conditions. Overall, the ratio of the 2X-glycosylated to the 1X-glycosylated to the aglycosylated mAb species under native conditions was approximately 88 to 5 to 7%, respectively. These results are comparable to ratios of major glycosylation states of the mAb observed under denaturing conditions (see Fig. 4C and Supporting Information Fig. 10).
4. Concluding remarks
In this study, an IgG type 1 mAb was analyzed by both middle-down and intact CZE-MS approaches. In the middle-down analysis using IdeS protease, with and without subsequent reduction, individual proteoforms were identified by a combination of CZE with single-stage MS analysis and CZE-MS2 with HCD fragmentation, enabling specific PTM site confirmations. Charge variants of the deamidated proteoforms were baseline separated by CZE. Different glycoforms of the FC/2 domain were also resolved. These results were complemented by intact mAb characterization, which revealed different populations of the mAb corresponding to 2X-glycosylated, 1X-glycosylated, and aglycosylated structures. Intact analysis revealed the presence of asymmetric glycans for the fully glycosylated protein, provided insights into the structural assembly of individual chains within the mAb, and allowed for quantitation of different intact proteoforms. These results were confirmed by the analysis of the mAb under native conditions, which also enabled the identification of dimers of the mAb.
Supplementary Material
Acknowledgments
We acknowledge the Biogen Analytical Development Department for their support through a Sponsored Research Agreement. This work was also supported by the National Institutes of Health under award numbers 1R01GM120272 and R01CA218500 (ARI). We acknowledge Thermo Fisher Scientific for their support through a technology alliance. The authors also thank SCIEX for providing CESI capillaries used in this study, the CESI 8000 instrument for CZE, and for insightful discussions.
Abbreviations:
- CESI
commercial sheathless interface
- cPEI
cross-linked polyethyleneimine
- CZE-MS
capillary zone electrophoresis – mass spectrometry
- HILIC
hydrophobic interaction chromatography
- IEX
ion exchange chromatography
- PTMs
post-translational modifications
- RPLC
reversed-phase liquid chromatography
Footnotes
The authors have declared no conflict of interest.
Additional supporting information may be found online in the Supporting Information section at the end of the article.
5 References
- [1].Elgundi Z, Reslan M, Cruz E, Sifniotis V, Kayser V, Adv. Drug Deliv. Rev 2017, 122, 2–19. [DOI] [PubMed] [Google Scholar]
- [2].Khawli LA, Goswami S, Hutchinson R, Kwong ZW, Yang J, Wang X, Yao Z, Sreedhara A, Cano T, Tesar D, Nijem I, Allison DE, Wong PY, Kao YH, Quan C, Joshi A, Harris RJ, Motchnik P, mAbs 2010, 2, 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Siuti N, Kelleher NL, Nat. Methods 2007, 4, 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fornelli L, Ayoub D, Aizikov K, Beck A, Tsybin YO, Anal. Chem 2014, 86, 3005–3012. [DOI] [PubMed] [Google Scholar]
- [5].Han M, Rock BM, Pearson JT, Rock DA, J. Chromatogr. B 2016, 1011, 24–32. [DOI] [PubMed] [Google Scholar]
- [6].Haselberg R, de Jong GJ, Somsen GW, Ana.l Chem 2013, 85, 2289–2296. [DOI] [PubMed] [Google Scholar]
- [7].Gahoual R, Beck A, Leize-Wagner E, Francois YN, J. Chromatogr. B Analyt. Technol. Biomed. Life Sci 2016, 1032, 61–78. [DOI] [PubMed] [Google Scholar]
- [8].Rosati S, Yang Y, Barendregt A, Heck AJR, Nat. Protocols 2014, 9, 967–976. [DOI] [PubMed] [Google Scholar]
- [9].Tran BQ, Barton C, Feng J, Sandjong A, Yoon SH, Awasthi S, Liang T, Khan MM, Kilgour DP, Goodlett DR, Goo YA, Data Brief 2016, 6, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dyachenko A, Wang G, Belov M, Makarov A, de Jong RN, van den Bremer ET, Parren PW, Heck AJ, Anal. Chem 2015, 87, 6095–6102. [DOI] [PubMed] [Google Scholar]
- [11].Bondarenko PV, Second TP, Zabrouskov V, Makarov AA, Zhang Z, J. Am. Soc. Mass Spectrom 2009, 20, 1415–1424. [DOI] [PubMed] [Google Scholar]
- [12].Kukrer B, Filipe V, van Duijn E, Kasper PT, Vreeken RJ, Heck AJR, Jiskoot W, Pharm. Res.-Dordr 2010, 27, 2197–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Muneeruddin K, Nazzaro M, Kaltashov IA, Anal. Chem 2015, 87, 10138–10145. [DOI] [PubMed] [Google Scholar]
- [14].Haverick M, Mengisen S, Shameem M, Ambrogelly A, mAbs 2014, 6, 852–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].D’Atri V, Fekete S, Beck A, Lauber MA, Guillarme D, Anal. Chem 2017, 89, 2086–2092. [DOI] [PubMed] [Google Scholar]
- [16].Dominguez-Vega E, Haselberg R, Somsen GW, Methods. Mol. Biol 2016, 1466, 25–41. [DOI] [PubMed] [Google Scholar]
- [17].Wojcik R, Dada OO, Sadilek M, Dovichi NJ, Rapid Commun. Mass Spectrom 2010, 24, 2554–2560. [DOI] [PubMed] [Google Scholar]
- [18].Redman EA, Mellors JS, Starkey JA, Ramsey JM, Anal. Chem 2016, 88, 2220–2226. [DOI] [PubMed] [Google Scholar]
- [19].Zhang B, Liu H, Karger BL, Foret F, Anal. Chem 1999, 71, 3258–3264. [DOI] [PubMed] [Google Scholar]
- [20].Busnel JM, Schoenmaker B, Ramautar R, Carrasco-Pancorbo A, Ratnayake C, Feitelson JS, Chapman JD, Deelder AM, Mayboroda OA, Anal. Chem 2010, 82, 9476–9483. [DOI] [PubMed] [Google Scholar]
- [21].Bush DR, Zang L, Belov AM, Ivanov AR, Karger BL, Anal. Chem 2016, 88, 1138–1146. [DOI] [PubMed] [Google Scholar]
- [22].Belov AM, Viner R, Santos MR, Horn DM, Bern M, Karger BL, Ivanov AR, J. Am. Soc. Mass Spectrom 2017, 28, 2614–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pontoglio A, Vigano A, Sebastiano R, Citterio A, Maragnoli L, Righetti PG, Gelfi C, Electrophoresis 2004, 25, 1065–1070. [DOI] [PubMed] [Google Scholar]
- [24].Verzola B, Sebastiano R, Righetti PG, Gelfi C, Lapadula M, Citterio A, Electrophoresis 2003, 24, 121–129. [DOI] [PubMed] [Google Scholar]
- [25].Batz NG, Mellors JS, Alarie JP, Ramsey JM, Anal. Chem 2014, 86, 3493–3500. [DOI] [PubMed] [Google Scholar]
- [26].Elhamili A, Wetterhall M, Arvidsson B, Sebastiano R, Righetti PG, Bergquist J, Electrophoresis 2008, 29, 1619–1625. [DOI] [PubMed] [Google Scholar]
- [27].Jensen PH, Karlsson NG, Kolarich D, Packer NH, Nat. Protoc 2012, 7, 1299–1310. [DOI] [PubMed] [Google Scholar]
- [28].Liu H, Gaza-Bulseco G, Faldu D, Chumsae C, Sun J, J. Pharm. Sci 2008, 97, 2426–2447. [DOI] [PubMed] [Google Scholar]
- [29].Quan C, Alcala E, Petkovska I, Matthews D, Canova-Davis E, Taticek R, Ma S, Anal. Biochem 2008, 373, 179–191. [DOI] [PubMed] [Google Scholar]
- [30].Zhang Q, Ames JM, Smith RD, Baynes JW, Metz TO, J. Proteome. Res 2009, 8, 754–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fu MX, Requena JR, Jenkins AJ, Lyons TJ, Baynes JW, Thorpe SR, J. Biol. Chem 1996, 271, 9982–9986. [DOI] [PubMed] [Google Scholar]
- [32].Liu H, May K, mAbs 2012, 4, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].McAuley A, Jacob J, Kolvenbach CG, Westland K, Lee HJ, Brych SR, Rehder D, Kleemann GR, Brems DN, Matsumura M, Protein Sci. 2008, 17, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Thies MJ, Talamo F, Mayer M, Bell S, Ruoppolo M, Marino G, Buchner J, J. Mol. Biol 2002, 319, 1267–1277. [DOI] [PubMed] [Google Scholar]
- [35].Isenman DE, Lancet D, Pecht I, Biochemistry 1979, 18, 3327–3336. [DOI] [PubMed] [Google Scholar]
- [36].Batra J, Rathore AS, Biotechnol. Prog 2016, 32, 1091–1102. [DOI] [PubMed] [Google Scholar]
- [37].Horne C, Klein M, Polidoulis I, Dorrington KJ, J. Immunol 1982, 129, 660–664. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.